Tenured Associate Professor
University of Florida J. Crayton Pruitt Family Department of Biomedical Engineering


Postdoc Position Open: Multiple fully funded Post-doc positions are open on research in Medical Artificial Intelligence (MAI), brain-inspired AI, and AI for precision brain health. Check out the "Join" tab to see more information.
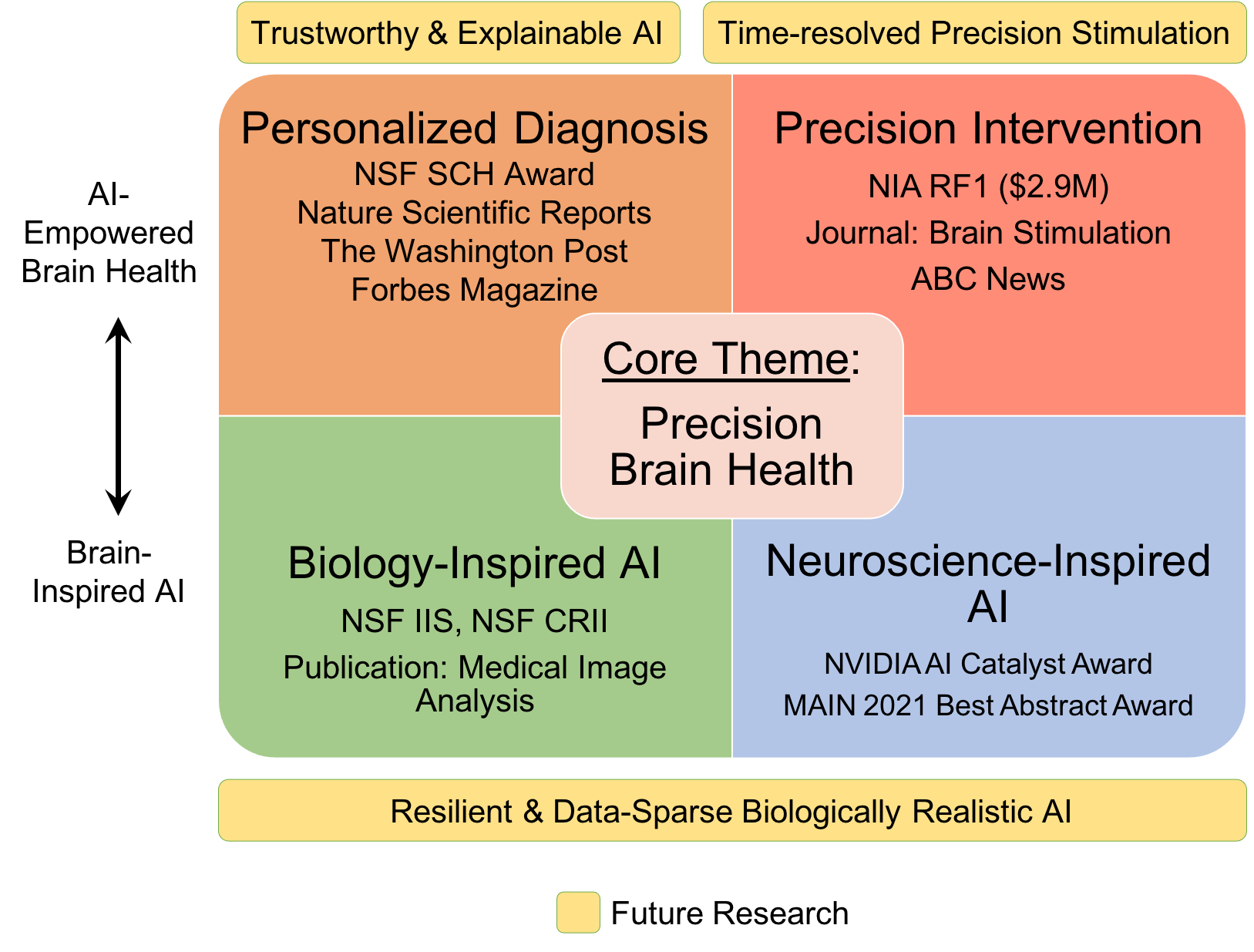
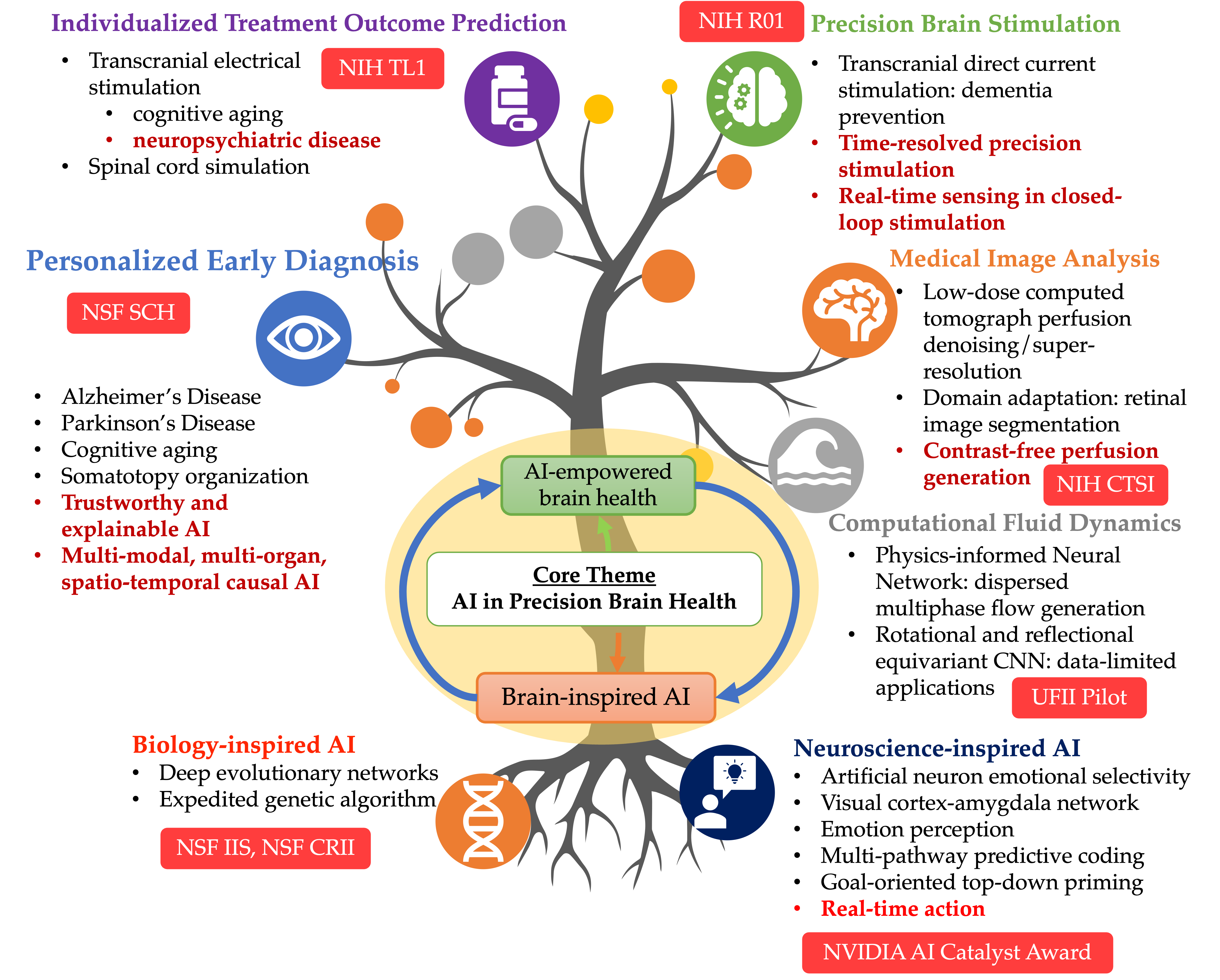
An AI researcher in medicine and healthcare, Dr. Ruogu Fang is a tenured Associate Professor and Pruitt Family Endowed Faculty Fellow in the J. Crayton Pruitt Family Department of Biomedical Engineering at the University of Florida. Her research encompasses two principal themes: AI-empowered precision brain health and brain/bio-inspired AI. Her work involves addressing compelling questions, such as using machine learning techniques to quantify brain dynamics, facilitating early Alzheimer's disease diagnosis through novel imagery, predicting personalized treatment outcomes, designing precision interventions, and leveraging principles from neuroscience to develop the next-generation of AI. Fang's current research is also rooted in the confluence of AI and multimodal medical image analysis. She is the PI of NIH NIA RF1 (R01-equivalent), NSF Research Initiation Initiative (CRII) Award, NSF CISE IIS Award, Ralph Lowe Junior Faculty Enhancement Award from Oak Ridge Associated Universities (ORAU). She has also received numerous recognitions. She was selected as the Rising Stars (Engineering) by the Academy of Science, Engineering, and Medicine of Florida (ASEMFL), inaugural recipient of the Robin Sidhu Memorial Young Scientist Award from the Society of Brain Mapping and Therapeutics, Best Paper Award from the IEEE International Conference on Image Processing, University of Florida Herbert Wertheim College of Engineering Faculty Award for Excellence in Innovation, UF BME Faculty Research Excellence Award, among others. Fang's research has been featured by Forbes Magazine, The Washington Post, ABC, RSNA, and published in Lancet Digital Health. She is an Associate Editor of the Journal Medical Image Analysis, a Topic Editor of Frontiers in Human Neuroscience, and a Guest Editor of CMGI. She is a reviewer for The Lancet, Nature Machine Intelligence, Science Advances, etc. Her research has been supported by NSF, NIH, Oak Ridge Laboratory, DHS, DoD, NVIDIA, and the University of Florida. She is President of Women in MICCAI (WiM) and Associate Editor of the Medical Image Analysis journal. At the heart of her work is the Smart Medical Informatics Learning and Evaluation (SMILE) lab, where she is tirelessly dedicated to creating groundbreaking brain and neuroscience-inspired medical AI and deep learning models. The primary objective of these models is to comprehend, diagnose, and treat brain disorders, all while navigating the complexities of extensive and intricate datasets.






Brain dynamics, which reflects the healthy or pathological states of the brain with quantifiable, reproducible, and indicative dynamics values, remains the least understood and studied area of brain science despite its intrinsic and critical importance to the brain. Unlike other brain information such as the structural and sequential dimensions that have all been extensively studied with models and methods successfully developed, the 5th dimension, dynamics, has only very recently started receiving systematic analysis from the research community. The state-of-the-art models suffer from several fundamental limitations that have critically inhibited the accuracy and reliability of the dynamic parameters' computation. First, dynamic parameters are derived from each voxel of the brain spatially independently, and thus miss the fundamental spatial information since the brain is connected? Second, current models rely solely on single-patient data to estimate the dynamic parameters without exploiting the big medical data consisting of billions of patients with similar diseases.
This project aims to develop a framework for data-driven brain dynamics characterization, modeling and evaluation that includes the new concept of a 5th dimension – brain dynamics – to complement the structural 4-D brain for a complete picture. The project studies how dynamic computing of the brain as a distinct problem from the image reconstruction and de-noising of convention models, and analyzes the impact of different models for the dynamics analysis. A data-driven, scalable framework will be developed to depict the functionality and dynamics of the brain. This framework enables full utilization of 4-D brain spatio-temporal data and big medical data, resulting in accurate estimations of the dynamics of the brain that are not reflected in the voxel-independent models and the single patient models. The model and framework will be evaluated on both simulated and real dual-dose computed tomography perfusion image data and then compared with the state-of-the-art methods for brain dynamics computation by leveraging collaborations with Florida International University Herbert Wertheim College of Medicine, NewYork-Presbyterian Hospital / Weill Cornell Medical College (WCMC) and Northwell School of Medicine at Hofstra University. The proposed research will significantly advance the state-of-the-art in quantifying and analyzing brain structure and dynamics, and the interplay between the two for brain disease diagnosis, including both the acute and chronic diseases. This unified approach brings together fields of Computer Science, Bioengineering, Cognitive Neuroscience and Neuroradiology to create a framework for precisely measuring and analyzing the 5th dimension – brain dynamics – integrated with the 4-D brain with three dimensions from spatial data and one dimension from temporal data. Results from the project will be incorporated into graduate-level multi-disciplinary courses in machine learning, computational neuroscience and medical image analysis. This project will open up several new research directions in the domain of brain analysis, and will educate and nurture young researchers, advance the involvement of underrepresented minorities in computer science research, and equip them with new insights, models and tools for developing future research in brain dynamics in a minority serving university.
Ph.D. in Electrical and Computer Engineering
Cornell University, Ithaca, NY.
B.E. in Information Engineering
Zhejiang University, Hangzhou, China
Women in MICCAI (WiM)
Medical Image Computing and Computer Assisted Intervention (MICCAI) Society
Medical Image Analysis
MICCAI Conference
The International Conference on Medical Image Computing and Computer Assited Intervention (MICCAI)
The Lancet, Nature Machine Intelligence, Science Advances, IEEE TPAMI, IEEE TNNLS, MIA, IEEE TMI, IEEE TIP
University of Florida J. Crayton Pruitt Family Department of Biomedical Engineering
University of Florida UF Intelligent Critical Care Center (IC3)
University of Florida Department of Electrical and Computer Engineering
University of Florida Department of Radiology, College of Medicine
University of Florida Department of Computer & Information Science & Engineering
University of Florida The Center for Cognitive Aging and Memory Clinical Translational Research (CAM)
University of Florida Genetics Institute (UFGI)
University of Florida UF Health Cancer Center
University of Florida Emerging Pathogens Institute























































































Human emotions are dynamic, multidimensional responses to challenges and opportunities that emerge from network interactions in the brain. Disruptions of these dynamics underlie emotional dysregulation in many mental disorders including anxiety and depression. To empirically study the neural basis of human emotion inference, experimenters often have observers view natural images varying in affective content, while at the same time recording their brain activity using electroencephalogram (EEG) and/or Functional magnetic resonance imaging (fMRI). Despite extensive research over the last few decades, much remains to be learned about the computational principles subserving the recognition of emotions in natural scenes. A major roadblock faced by empirical neuroscientists is the inability to carry out precisely manipulate human neural systems and test the consequences in imaging data. Deep Neural Networks (DNN), owing to their high relevance to human neural systems and extraordinary prediction capability, have become a promising tool for testing these sorts of hypotheses in swift and nearly costless computer simulations. The overarching goal of this project is to develop a neuroscience-inspired, DNN-based deep learning framework for emotion inference in real-world scenarios by synergistically integrating neuron-, circuit-, and system-level mechanisms. Recognizing that the state-of-the-art DNNs are centered on bottom-up and feedforward-only processing, which disagrees with the strong goal-oriented top-down modulation recurrence observed in the physiology, this project aims to enrich DNNs and enable closer AI-neuroscience interaction by incorporating goal-oriented top-down modulation and reciprocal interactions DNNs and test the model assumptions and predictions on neuroimaging data. To meet these goals, the project aims to develop a brain-inspired goal-oriented and bidirectional deep learning model for emotion inference. Despite the great promise shown by today?s deep learning as a framework for modeling biological vision, their architecture is limited to emulating the visual cortex for face/object/scene recognition and rarely goes beyond the inferotemporal cortex (IT), which is necessary for modeling high-level cognitive processes. In this project, we propose to build a biologically plausible deep learning architecture by integrating an in-silico amygdala module into the visual cortex architecture in DNN (the VCA model). The researchers hope to build neuron-, circuit-, and system-level modulation via goal-oriented attention priming, and multi-pathway predictive coding to 1) elucidate the mechanism of selectivity underlying preference and response to naturalistic emotions by artificial neurons; 2) differentiate fine-grained emotional responses via multi-path predictive coding, and 3) refine the neuroscientific understanding of human neuro-behavioral data by comparing attention priming and temporal generalization observed in simultaneous fMRI-EEG data to the computational observations using our brain-inspired VCA model. This project introduces two key innovations, both patterned after how brain operates, into DNN architecture and demonstrate their superior performance when applied to complex real-world tasks. Successful execution of the project can lead to the development of a new generation of AI-models that are inspired by neuroscience and that may in turn power neuroscience research.

There is a pressing need for effective interventions to remediate age-related cognitive decline and alter the trajectory toward Alzheimer’s disease. The NIA Alzheimer’s Disease Initiative funded Phase III Augmenting Cognitive Training in Older Adults (ACT) trial aimed to demonstrate that transcranial direct current stimulation (tDCS) paired with cognitive training could achieve this goal. The present study proposes a state of the art secondary data analysis of ACT trial data that will further this aim by 1) elucidate mechanism of action underlying response to tDCS treatment with CT, 2) address heterogeneity of response in tDCS augmented CT by determining how individual variation in the dose of electrical current delivered to the brain interacts with individual brain anatomical characteristics; and 3) refine the intervention strategy of tDCS paired with CT by evaluating methods for precision delivery targeted dosing characteristics to facilitate tDCS augmented outcomes. tDCS intervention to date, including ACT, apply a fixed dosing approach whereby a single stimulation intensity (e.g., 2mA) and set of electrode positions on the scalp (e.g., F3/F4) is applied to all participants/patients. However, our recent work has demonstrated that age-related changes in neuroanatomy as well as individual variability in head/brain structures (e.g., skull thickness) significantly impacts the distribution and intensity of electrical current induced in the brain from tDCS. This project will use person-specific MRI-derived finite element computational models of electric current characteristics (current intensity and direction of current flow) and new methods for enhancing the precision and accuracy of derived models to precisely quantify the heterogeneity of current delivery in older adults. We will leverage these individualized precision models with state-of-the-art support vector machine learning methods to determine the relationship between current characteristics and treatment response to tDCS and CT. We will leverage the inherent heterogeneity of neuroanatomy and fixed current delivery to provide insight in the not only which dosing parameters were associated with treatment response, but also brain region specific information to facilitate targeted delivery of stimulation in future trials. Further still, the current study will also pioneer new methods for calculation of precision dosing parameters for tDCS delivery to potentially optimize treatment response, as well as identify clinical and demographic characteristics that are associated with response to tDCS and CT in older adults. Leveraging a robust and comprehensive behavioral and multimodal neuroimaging data set for ACT with advanced computational methods, the proposed study will provide critical information for mechanism, heterogeneity of treatment response and a pathway to refined precision dosing approaches for remediating age- related cognitive decline and altering the trajectory of older adults toward Alzheimer’s disease.

There is a pressing need for effective interventions to remediate age-related cognitive decline and alter the trajectory toward Alzheimer’s disease. The NIA Alzheimer’s Disease Initiative funded Phase III Augmenting Cognitive Training in Older Adults (ACT) trial aimed to demonstrate that transcranial direct current stimulation (tDCS) paired with cognitive training could achieve this goal. The present study proposes a state of the art secondary data analysis of ACT trial data that will further this aim by 1) elucidate mechanism of action underlying response to tDCS treatment with CT, 2) address heterogeneity of response in tDCS augmented CT by determining how individual variation in the dose of electrical current delivered to the brain interacts with individual brain anatomical characteristics; and 3) refine the intervention strategy of tDCS paired with CT by evaluating methods for precision delivery targeted dosing characteristics to facilitate tDCS augmented outcomes. tDCS intervention to date, including ACT, apply a fixed dosing approach whereby a single stimulation intensity (e.g., 2mA) and set of electrode positions on the scalp (e.g., F3/F4) is applied to all participants/patients. However, our recent work has demonstrated that age-related changes in neuroanatomy as well as individual variability in head/brain structures (e.g., skull thickness) significantly impacts the distribution and intensity of electrical current induced in the brain from tDCS. This project will use person-specific MRI-derived finite element computational models of electric current characteristics (current intensity and direction of current flow) and new methods for enhancing the precision and accuracy of derived models to precisely quantify the heterogeneity of current delivery in older adults. We will leverage these individualized precision models with state-of-the-art support vector machine learning methods to determine the relationship between current characteristics and treatment response to tDCS and CT. We will leverage the inherent heterogeneity of neuroanatomy and fixed current delivery to provide insight in the not only which dosing parameters were associated with treatment response, but also brain region specific information to facilitate targeted delivery of stimulation in future trials. Further still, the current study will also pioneer new methods for calculation of precision dosing parameters for tDCS delivery to potentially optimize treatment response, as well as identify clinical and demographic characteristics that are associated with response to tDCS and CT in older adults. Leveraging a robust and comprehensive behavioral and multimodal neuroimaging data set for ACT with advanced computational methods, the proposed study will provide critical information for mechanism, heterogeneity of treatment response and a pathway to refined precision dosing approaches for remediating age- related cognitive decline and altering the trajectory of older adults toward Alzheimer’s disease.

Driven by its performance accuracy, machine learning (ML) has been used extensively for various applications in the healthcare domain. Despite its promising performance, researchers and the public have grown alarmed by two unsettling deficiencies of these otherwise useful and powerful models. First, there is a lack of trustworthiness - ML models are prone to interference or deception and exhibit erratic behaviors when in action dealing with unseen data, despite good practice during the training phase. Second, there is a lack of interpretability - ML models have been described as 'black-boxes' because there is little explanation for why the models make the predictions they do. This has called into question the applicability of ML to decision-making in critical scenarios such as image-based disease diagnostics or medical treatment recommendation. The ultimate goal of this project is to develop computational foundation for trustworthy and explainable Artificial Intelligence (AI), and offer a low-cost and non-invasive ML-based approach to early diagnosis of neurodegenerative diseases. In particular, the project aims to develop computational theories, ML algorithms, and prototype systems. The project includes developing principled solutions to trustworthy ML and making the ML prediction process transparent to end-users. The later will focus on explaining how and why an ML model makes such a prediction, while dissecting its underlying structure for deeper understanding. The proposed models are further extended to a multi-modal and spatial-temporal framework, an important aspect of applying ML models to healthcare. A verification framework with end-users is defined, which will further enhance the trustworthiness of the prototype systems. This project will benefit a variety of high-impact AI-based applications in terms of their explainability, trustworthy, and verifiability. It not only advances the research fronts of deep learning and AI, but also supports transformations in diagnosing neurodegenerative diseases.
This project will develop the computational foundation for trustworthy and explainable AI with several innovations. First, the project will systematically study the trustworthiness of ML systems. This will be measured by novel metrics such as, adversarial robustness and semantic saliency, and will be carried out to establish the theoretical basis and practical limits of trustworthiness of ML algorithms. Second, the project provides a paradigm shift for explainable AI, explaining how and why a ML model makes its prediction, moving away from ad-hoc explanations (i.e. what features are important to the prediction). A proof-based approach, which probes all the hidden layers of a given model to identify critical layers and neurons involved in a prediction from a local point of view, will be devised. Third, a verification framework, where users can verify the model's performance and explanations with proofs, will be designed to further enhance the trustworthiness of the system. Finally, the project also advances the frontier of neurodegenerative diseases early diagnosis from multimodal imaging and longitudinal data by: (i) identifying retinal vasculature biomarkers using proof-based probing in biomarker graph networks; (ii) connecting biomarkers of the retina and the brain vasculature via cross- modality explainable AI model; and, (iii) recognizing the longitudinal trajectory of vasculature biomarkers via a spatio-temporal recurrent explainable model. This synergistic effort between computer science and medicine will enable a wide range of applications to trustworthy and explainable AI for healthcare. The results of this project will be assimilated into the courses and summer programs that the research team have developed with specially designed projects to train students with trustworthy and explainable AI.

History has taught us that exposures to radionuclides can happen any day almost anywhere in the US and elsewhere and we have done little to prepare ourselves. Our ability to perform dosimetry modeling for such scenarios and efforts into biomarker and mitigation discovery are archaic and our tendency to rely on external beam radiation to model these is utterly misplaced. We should and we can do much better. This program centers on the hypothesis that radiation from internal emitters is very unevenly distributed within a body, amongst organs, and even within organs, tissues and cells. The half-life and decay schema of the radionuclide, its activity and concentration, particle size and morphology, and its chemical form and solubility are all critical, as are the route of uptake, tissue structure, genetic makeup, physiology, danger signaling and the crosstalk with the immune system. Conceptually this suggests that the analysis of radionuclide distribution requires measurements at the MESO, MICRO and NANO level for accurate dosimetry modeling and biokinetics analyses, that will much better align with biological endpoints, and therefore with meaningful countermeasure development. In many ways our program integrates the three main pillars of radiation science, namely radiation physics, radiation chemistry and radiation biology, taking into account pharmacokinetics and pharmacodynamics aspects of particle distribution at subcellular, cellular, and tissue levels. In other words, to understand the biological effects of internal emitters and find the best possible mitigation strategies a systematic study is called for, one that includes but is not limited to: a) radionuclide physical and chemical form and intravital migration, b) protracted exposure times, c) radiation quality parameters, d) novel virtual phantom modeling beyond few MACRO reference models ; e) novel biokinetics with sex- and age- specificity; f) MESO, MICRO and NANO scale histology and immunohistochemistry with integrated radionuclide distribution information; g) exploration of molecular biomarkers of radionuclide intake and contamination and h) countermeasures that modulate radionuclide distribution and possibly also improve DNA, cell and tissue repair. We have assembled a team with diverse scientific expertise that can tackle these challenges within an integrated program. There is an incredibly impressive technological toolbox at our disposal and our goal is to generate a meaningful blueprint for understanding and predicting biological consequences of exposure to radionuclides. The possible benefits of this program to the radiation research community and the general population are immense.

The purpose of the SHARE P01 research program project is to address HIV and alcohol use around three themes; 1) Emerging adulthood (ages 18 -29); 2) Self-management of HIV and alcohol; and 3) Translational behavioral science. Emerging adulthood is a developmental stage marked by significant change in social roles, expectations as a new adult, and increased responsibilities. It is also marked by poor HIV self- management and increased alcohol use. Emerging adults with HIV (hereafter called young people living with HIV; YPLWH) may face even more challenges given intersectional stigma. This age group continues to have very high rates of new HIV infections. Interventions designed specifically for the unique developmental challenges of emerging adults are needed, yet emerging adults are often included with older adults in intervention programs. The concept of self-management emerged concurrently within both the substance abuse and chronic illness literatures, and fits well with the developmental challenges of emerging adulthood. Self-management, a framework we have utilized in our work with YPLWH, refers to the ability to manage symptoms, treatments, lifestyle changes, and consequences of health conditions. Current research now identifies individual-level self-management skills such as self-control, decision-making, self-reinforcement, and problem solving as that protect against substance use and improve other health outcomes and can be embedded in the Information-Motivation-Behavioral Skills model. Although we have conducted multiple studies with YPLWH, only one intervention to date (Healthy Choices conducted by our team) improved both alcohol use and viral suppression in YPLWH in large trials. The goal of the SHARE P01 is to utilize advances in translational behavioral science to optimize behavioral interventions and define new developmentally- and culturally-appropriate intervention targets to improve self-management of alcohol and HIV in YPLWH. We will focus our efforts in Florida, a state hardest hit by the HIV epidemic but with a particularly strong academic- community partnership to support translation. We have assembled research teams to conduct self- management studies across the translational spectrum to address self-management and improve alcohol use and viral suppression (and thereby reduce transmission) in diverse YPLWH in Florida. The P01 will consist of three research projects (DEFINE, ENGAGE, and SUSTAIN), representing different stages on the translational spectrum and targeting different core competencies, supported by two cores (Community Engagement Core and Data Science Core). If successful, the SHARE P01 has the potential to greatly advance programs promoting self-management of HIV and alcohol use among a particularly vulnerable, but under-researched group, emerging adults living with HIV. SHARE also has a high potential for scale-up and implementation beyond Florida and across the United States.

As persons living with HIV (PLWH) live longer, approximately 50% will experience HIV-related cognitive dysfunction, which may affect daily activities, contribute to morbidity and mortality, and increase the likelihood of HIV transmission. Alcohol consumption among PLWH may further exacerbate long-term cognitive dysfunction, with the presumed mechanism involving the gut microbiome, microbial translocation, systemic inflammation, and ultimately neuroinflammation. However, there are many gaps in our understanding regarding the specific pathophysiological mechanisms, and a need to offer interventions that are effective and acceptable in helping PLWH to reduce drinking or to protect them against alcohol-related harm. The overarching goal of this P01 is to identify and ultimately implement new/improved, targeted interventions that will improve outcomes related to cognitive and brain dysfunction in persons with HIV who drink alcohol. The proposed P01 activity will extend our current line of research that forms the core of the Southern HIV & Alcohol Research Consortium (SHARC). The specific aims of this P01 are to: 1) improve our understanding of the specific mechanisms that connect the gut microbiome to cognitive and brain health outcomes in persons with HIV; 2) evaluate interventions that are intended to reduce the impact of alcohol on brain and cognitive health in persons with HIV; and 3) connect and extend the research activity from this P01 with the training programs and community engagement activity in the SHARC. Our P01 will utilize two cores that provide infrastructure to two Research Components (RC1, RC2). The two RC will together enroll 200 PLWH with at-risk drinking into clinical trials that share common timepoints and outcome assessments. RC1 will compare two strategies to extend contingency management to 60 days, using breathalyzers and wrist-worn biosensors to monitor drinking. RC2 uses a hybrid trial design to evaluate two biomedical interventions targeting the gut-brain axis. One intervention is a wearable, transcutaneous vagus nerve stimulator that is hypothesized to stimulate the autonomic nervous system, resulting in decreased inflammation and improved cognition. The other intervention is a probiotic supplement intended to improve the gut microbiome in persons with HIV and alcohol consumption. All participants in RC2, and a subset of those in RC1 will have neuroimaging at two timepoints. The Data Science Core will provide data management and analytical support, and will analyze existing data and the data collected from this P01 using a machine learning and AI approach to identify factors associated with intervention success or failure. The Administrative Core will provide scientific leadership, clinical research and recruitment infrastructure, and connection to the outstanding training programs, development opportunities, and community engagement provided by the SHARC. Our community engagement with diverse populations, and collection of acceptability data from clinical trial participants, will facilitate our readiness to scale up the most promising interventions and move towards implementation in the next phase of our research.

The number of persons living with HIV (PLWH) continues to increase in the United States. Alcohol consumption is a significant barrier to both achieving the goal of ending the HIV epidemic and preventing comorbidities among PLWH, as it contributes to both HIV transmission and HIV-related complications. Recent advances in data capture systems such as mHealth devices, medical imaging, and high-throughput biotechnologies make large/complex research and clinical datasets available, including survey data, multi-omics data, electronic medical records, and/or other sources of reliable information related to engagement in care. This offers tremendous potential of applying “big” data to extract knowledge and insights regarding fundamental physiology, understand the mechanisms by which the pathogenic effects of biotic and abiotic factors are realized, and identify potential intervention targets. We propose to integrate the disparate data sources maintained by our partners and then utilize the big data to address research questions in treating HIV and alcohol-related morbidity and mortality. Specifically, we will pursue the following three aims: 1) Integrate the disparate data sources through standardization, harmonization, and merging; 2) Develop a web-based data sharing platform including virtual data sharing communities, data privacy protection, streamlined data approval and access, and tracking of ongoing research activities; 3) Provide statistical support to junior investigators to use the data repository for exploratory data analysis and proposal development. The proposed study will tap into disparate data sources, unleash the potential of data and information, accelerate knowledge discovery, advance data-powered health, and transform discovery to improve health outcomes for PLWH.

Classical aversive conditioning is a well-established laboratory model for studying acquisition and extinction of defensive responses. In experimental animals, as well as in humans, research to date has been mainly focused on the role of limbic structures (e.g., the amygdala) in these responses. Recent evidence has begun to stress the important contribution by the brain’s sensory and attention control systems in maintaining the neural representations of conditioned responses and in facilitating their extinction. The proposed research breaks new ground by combining novel neuroimaging techniques with advanced computational methods to examine the brain’s visual and attention processes underlying fear acquisition and extinction in humans. Major advances will be made along three specific aims. In Aim 1, we characterize the brain network dynamics of visuocortical threat bias formation, extinction, and recall in a two-day learning paradigm. In Aim 2, we establish and test a computational model of threat bias generalization. In Aim 3, we examine the relation between individual differences in generalization and recall of conditioned visuocortical threat biases and individual differences in heightened autonomic reactivity to conditioned threat, a potential biomarker for assessing the predisposition to developing the disorders of fear and anxiety. It is expected that accomplishing these research aims will address two NIMH strategic priorities: defining the circuitry and brain networks underlying complex behaviors (Objective 1) and identifying and validating new targets for treatment that are derived from the understanding of disease mechanisms (Objective 3). It is further expected that this project will enable a paradigm shift in research on dysfunctional attention to threat from one that focuses primarily on limbic-prefrontal circuits to one that emphasizes the interactions among sensory, attention, executive control and limbic systems.

Across the globe, there has been a considerable growth in the number of people diagnosed with Parkinsonism. Estimates indicate that from 1990 to 2015 the number of Parkinsonism diagnoses doubled, with more than 6 million people currently carrying the diagnosis, and by year 2040, 12 and 14.2 million people will be diagnosed with Parkinsonism. Parkinson’s disease (PD), multiple system atrophy Parkinsonian variant (MSAp), and progressive supranuclear palsy (PSP) are neurodegenerative forms of Parkinsonism, which can be difficult to diagnose as they share similar motor and non-motor features, and they each have an increased chance of developing dementia. In the first five years of a PD diagnosis, about 58% of PD are misdiagnosed, and of these misdiagnoses about half have either MSA or PSP. Since PD, MSAp, and PSP require unique treatment plans and different medications, and clinical trials testing new medications require the correct diagnosis, there is an urgent need for both clinic ready and clinical-trial ready markers for differential diagnosis of PD, MSAp, and PSP. Over the past decade, we have developed diffusion imaging as an innovative biomarker for differentiating PD, MSAp, and PSP. In this proposal, we will leverage our extensive experience to create a web-based software tool that can process diffusion imaging data from anywhere in the world. We will disseminate and test the tool in the largest prospective cohort of participants with Parkinsonism (PD, MSAp, PSP), working closely with the Parkinson Study Group. The reason to test this in the Parkinson Study Group network, is because they are the community that evaluates Phase II and Phase III clinical trials in Parkinsonism. This web-based software tool will be capable of reading raw diffusion imaging data, performing quality assurance procedures, analyzing the data using a validated pipeline, and providing imaging metrics and diagnostic probability. We will test the performance of the wAID-P by enrolling 315 total subjects (105 PD, 105 MSAp, 105 PSP) across 21 sites in the Parkinson Study Group. Each site will perform imaging, clinical scales, diagnosis, and will upload the data to the web-based software tool. The clinical diagnosis will be blinded to the diagnostic algorithm and the imaging diagnosis will be compared to the movement disorders trained neurologist diagnosis. We will also enroll a portion of the cohort into a brain bank to ascertain pathological confirmation and to test the algorithm against cases with post-mortem diagnoses. The final outcome will be to disseminate a validated diagnostic algorithm to the Parkinson neurological and radiological community and to make it available to all on a website.

The temporal dynamics of blood flows through the network of cerebral arteries and veins provides a window into the health of the human brain. Since the brain is vulnerable to disrupted blood supply, brain dynamics serves as a crucial indicator for many kinds of neurological diseases such as stroke, brain cancer, and Alzheimer's disease. Existing efforts at characterizing brain dynamics have predominantly centered on 'isolated' models in which data from single-voxel, single-modality, and single-subject are characterized. However, the brain is a vast network, naturally connected on structural and functional levels, and multimodal imaging provides complementary information on this natural connectivity. Thus, the current isolated models are deemed not capable of offering the platform necessary to enable many of the potential advancements in understanding, diagnosing, and treating neurological and cognitive diseases, leaving a critical gap between the current computational modeling capabilities and the needs in brain dynamics analysis. This project aims to bridge this gap by exploiting multi-scale structural (voxel, vasculature, tissue) connectivity and multi-modal (anatomical, angiography, perfusion) connectivity to develop an integrated connective computational paradigm for characterizing and understanding brain dynamics.

The temporal dynamics of blood flows through the network of cerebral arteries and veins provides a window into the health of the human brain. Since the brain is vulnerable to disrupted blood supply, brain dynamics serves as a crucial indicator for many kinds of neurological diseases such as stroke, brain cancer, and Alzheimer's disease. Existing efforts at characterizing brain dynamics have predominantly centered on 'isolated' models in which data from single-voxel, single-modality, and single-subject are characterized. However, the brain is a vast network, naturally connected on structural and functional levels, and multimodal imaging provides complementary information on this natural connectivity. Thus, the current isolated models are deemed not capable of offering the platform necessary to enable many of the potential advancements in understanding, diagnosing, and treating neurological and cognitive diseases, leaving a critical gap between the current computational modeling capabilities and the needs in brain dynamics analysis. This project aims to bridge this gap by exploiting multi-scale structural (voxel, vasculature, tissue) connectivity and multi-modal (anatomical, angiography, perfusion) connectivity to develop an integrated connective computational paradigm for characterizing and understanding brain dynamics.

This project establishes the NSF Industry/University Collaborative Research Center for Big Learning (CBL). The vision is to create intelligence towards intelligence-driven society. Through catalyzing the fusion of diverse expertise from the consortium of faculty members, students, industry partners, and federal agencies, CBL seeks to create state-of-the-art deep learning methodologies and technologies and enable intelligent applications, transforming broad domains, such as business, healthcare, Internet-of-Things, and cybersecurity. This timely initiative creates a unique platform for empowering our next-generation talents with cutting-edge technologies of societal relevance and significance. This project establishes the NSF Industry/University Collaborative Research Center for Big Learning (CBL) at University of Florida (UF). With substantial breakthroughs in multiple modalities of challenges, such as computer vision, speech recognitions, and natural language understanding, the renaissance of machine intelligence is dawning. The CBL vision is to create intelligence towards intelligence-driven society. The mission is to pioneer novel deep learning algorithms, systems, and applications through unified and coordinated efforts in the CBL consortium. The UF Site will focus on intelligent platforms and applications and closely collaborate with other sites on deep learning algorithms, systems, and applications. The CBL will have broad transformative impacts in technologies, education, and society. CBL aims to create pioneering research and applications to address a broad spectrum of real-world challenges, making significant contributions and impacts to the deep learning community. The discoveries from CBL will make significant contributions to promote products and services of industry in general and CBL industry partners in particular. As the magnet of deep learning research and applications, CBL offers an ideal platform to nurture next-generation talents through world-class mentors from both academia and industry, disseminates the cutting-edge technologies, and facilitates industry/university collaboration and technology transfer. The center repository will be hosted at http://nsfcbl.org. The data, code, documents will be well organized and maintained on the CBL servers for the duration of the center for more than five years and beyond. The internal code repository will be managed by GitLab. After the software packages are well documented and tested, they will be released and managed by popular public code hosting services, such as GitHub and Bitbucket.

Human emotions are dynamic, multidimensional responses to challenges and opportunities, which emerge from network interactions in the brain. Disruptions of these network interactions underlie emotional dysregulation in many mental disorders, including anxiety and depression. Creating an AI-based model system that is informed and validated by known biological findings and can be used to carry out causal manipulations and test the consequences against human imaging data will thus be a highly significant development in the short term. The long-term goal is to understand how the human brain processes emotional information and how the process breaks down in mental disorders. NIH currently funds the team to record and analyze fMRI data from humans viewing natural images of varying emotional content. In the process of their research, they recognize that empirical studies such as theirs have significant limitations. Chief among them is the lack of ability to manipulate the system to establish the causal basis for the observed relationship between brain and behavior. The advent of AI, especially deep neural networks (DNNs), opens a new avenue to address this problem. Creating an AI-based model system that is informed and validated by known biological findings and that can be used to carry out causal manipulations and allow the testing of the consequences against human imaging data will thus be a significant step toward achieving our long-term goal.

Human emotions are dynamic, multidimensional responses to challenges and opportunities, which emerge from network interactions in the brain. Disruptions of these network interactions underlie emotional dysregulation in many mental disorders, including anxiety and depression. Creating an AI-based model system that is informed and validated by known biological findings and can be used to carry out causal manipulations and test the consequences against human imaging data will thus be a highly significant development in the short term. The long-term goal is to understand how the human brain processes emotional information and how the process breaks down in mental disorders. NIH currently funds the team to record and analyze fMRI data from humans viewing natural images of varying emotional content. In the process of their research, they recognize that empirical studies such as theirs have significant limitations. Chief among them is the lack of ability to manipulate the system to establish the causal basis for the observed relationship between brain and behavior. The advent of AI, especially deep neural networks (DNNs), opens a new avenue to address this problem. Creating an AI-based model system that is informed and validated by known biological findings and that can be used to carry out causal manipulations and allow the testing of the consequences against human imaging data will thus be a significant step toward achieving our long-term goal.

Vision-threatening diseases are one of the leading causes of blindness. DR, a common complication of diabetes, is the leading cause of blindness in American adults and the fastest growing disease threatening nearly 415 million diabetic patients worldwide. With professional eye imaging devices such as fundus cameras or Optical Coherence Tomography (OCT) scanners, most of the vision-threatening diseases can be curable if detected early. However, these diseases are still damaging people’s vision and leading to irreversible blindness, especially in rural areas and low-income communities where professional imaging devices and medical specialists are not available or not even affordable. There is an urgent need for early detection of vision-threatening diseases before vision loss in these areas. The current practice of DR screening relies on human experts to manually examine and diagnose DR in stereoscopic color fundus photographs at hospitals using professional fundus camera, which is time-consuming and infeasible for large-scale screening. It also puts an enormous burden on ophthalmologists and increases waiting lists and may undermine the standards of health care. Therein, automatic DR diagnosis systems with ophthalmologist-level performance are a critical and unmet need for DR screening. Electronic Health Records (EHR) have been increasingly implemented in US hospitals. Vast amounts of longitudinal patient data have been accumulated and are available electronically in structured tables, narrative text, and images. There is an increasing need for multimodal synergistic learning methods to link different data sources for clinical and translational studies. Recent emerging of AI technologies, especially deep learning (DL) algorithms, have greatly improved the performance of automated vision-disease diagnosis systems based on EHR data. However, the current systems are unable to detect early stage of vision-diseases. On the other hand, the clinical text provides detailed diagnosis, symptoms, and other critical observations documented by physicians, which could be a valuable resource to help lesion detection from medical images. Multimodal synergistic learning is the key to linking clinical text to medical images for lesion detection. This study proposes to leverage the narrative clinical text to improve lesion level detection from medical images via clinical Natural Language Processing (NLP). The team hypothesizes that early stage vision-threatening diseases can be detected using smartphone-based fundus camera via multimodal learning integrating clinical text and images with limited lesion-level labels via clinical NLP. The ultimate goal is to improve the early detection and prevention of vision-threatening diseases among rural and low-income areas by developing a low-cost, highly efficient system that can leverage both clinical narratives and images.




In the current era of invigorating brain research, there is emerging attention in leveraging machine learning in understanding the brain, particularly exploring the brain dynamics. With the ever-increasing amount of neuroscience data, new challenges and opportunities arise for brain dynamics analysis, such as data-driven reconstruction and computer-aided diagnosis. However, there are few attempts to bridge the semantic gaps between the raw brain imaging data and the diagnosis. We will develop robust and data-driven techniques for the purpose of modeling, estimating functional parameters from the limited data brain images, and making decision support practical based on efficient direct estimation of the brain dynamics. This is an interdisciplinary research combining medical image analysis, machine learning, neuroscience, and the domain expertise.

In the midst of an outbreak or sustained epidemic, reliable prediction of transmission risks and patterns of spread is critical to inform public health programs. Projections of transmission growth or decline among specific risk groups can aid in optimizing interventions, particularly when resources are limited. Phylogenetic trees have been widely used in the detection of transmission chains and high-risk populations. Moreover, tree topology and the incorporation of population parameters (phylodynamics) can be useful in reconstructing the evolutionary dynamics of an epidemic across space and time among individuals. We now demonstrate the utility of phylodynamic trees for transmission modeling and forecasting, developing a phylogeny-based deep learning system, referred to as \textit{DeepDynaForecast}. Our approach leverages a primal-dual graph learning structure with shortcut multi-layer aggregation, which is suited for the early identification and prediction of transmission dynamics in emerging high-risk groups. We demonstrate the accuracy of \textit{DeepDynaForecast} using simulated outbreak data and the utility of the learned model using empirical, large-scale data from the human immunodeficiency virus epidemic in Florida between 2012 and 2020. Our framework is available as open-source software (MIT license) at github.com/lab-smile/DeepDynaForcast.

Alzheimer’s Disease (AD) is a progressive neurodegenerative disease and the leading cause of dementia. Early diagnosis is critical for patients to benefit from potential intervention and treatment. The retina has emerged as a plausible diagnostic site for AD detection owing to its anatomical connection with the brain. However, existing AI models for this purpose have yet to provide a rational explanation behind their decisions and have not been able to infer the stage of the disease’s progression. Along this direction, we propose a novel model-agnostic explainable-AI framework, called Granur Neuron-leel Expliner (LAVA), an interpretation prototype that probes into intermediate layers of the Convolutional Neural Network (CNN) models to directly assess the continuum of AD from the retinal imaging without the need for longitudinal or clinical evaluations. This innovative approach aims to validate retinal vasculature as a biomarker and diagnostic modality for evaluating Alzheimer’s Disease. Leveraged UK Biobank cognitive tests and vascular morphological features demonstrate significant promise and effectiveness of LAVA in identifying AD stages across the progression continuum.
Recent neuroimaging studies have shown that the visual cortex plays an important role in representing the affective significance of visual input. The origin of these affect-specific visual representations is debated: they are intrinsic to the visual system versus they arise through reentry from frontal emotion processing structures such as the amygdala. We examined this problem by combining convolutional neural network (CNN) models of the human ventral visual cortex pre-trained on ImageNet with two datasets of affective images. Our results show that in all layers of the CNN models, there were artificial neurons that responded consistently and selectively to neutral, pleasant, or unpleasant images and lesioning these neurons by setting their output to zero or enhancing these neurons by increasing their gain led to decreased or increased emotion recognition performance respectively. These results support the idea that the visual system may have the intrinsic ability to represent the affective significance of visual input and suggest that CNNs offer a fruitful platform for testing neuroscientific theories.

Improving the diagnosis of HIV is a fundamental objective of the Ending the HIV Epidemic initiative, as it represents the initial step toward treatment and achieving undetectable status, thereby reducing transmission. To attain these objectives effectively, it is crucial to identify the groups most susceptible to HIV, allowing interventions to be tailored to their specific needs. In this study, we developed a predictive model designed to assess individual HIV risk within a high-risk contact network predicting treatment or at-risk leveraging surveillance data collected through routine HIV case interviews in Florida. Unique to our analysis, we explored the incorporation of behavioral network information with Graph Neural Networks to enhance the predictive capacity for identifying individuals within the treatment or intervention categories, when compared to models that mainly consider conventional HIV risk factors. Our deployed Graph Isomorphism Network achieved 77.3% and 73.2% balanced accuracy in inductive and transductive learning scenarios respectively, outperforming the traditional prediction algorithms that do not leverage the network structure. We then used our model to further investigate the importance of demographic and behavioral factors in the HIV risk prediction process. Our findings provide valuable insights for healthcare practitioners and policymakers in their efforts to combat HIV infection.

Parkinson’s disease is the world’s fastest-growing neurological disorder. Research to elucidate the mechanisms of Parkinson’s disease and automate diagnostics would greatly improve the treatment of patients with Parkinson’s disease. Current diagnostic methods are expensive and have limited availability. Considering the insidious and preclinical onset and progression of the disease, a desirable screening should be diagnostically accurate even before the onset of symptoms to allow medical interventions. We highlight retinal fundus imaging, often termed a window to the brain, as a diagnostic screening modality for Parkinson’s disease. We conducted a systematic evaluation of conventional machine learning and deep learning techniques to classify Parkinson’s disease from UK Biobank fundus imaging. Our results suggest Parkinson’s disease individuals can be differentiated from age and gender-matched healthy subjects with 68% accuracy. This accuracy is maintained when predicting either prevalent or incident Parkinson’s disease. Explainability and trustworthiness are enhanced by visual attribution maps of localized biomarkers and quantified metrics of model robustness to data perturbations.

Whole-head segmentation from Magnetic Resonance Images (MRI) establishes the foundation for individualized computational models using finite element method (FEM). This foundation paves the path for computer-aided solutions in fields, particularly in non-invasive brain stimulation. Most current automatic head segmentation tools are developed using healthy young adults. Thus, they may neglect the older population that is more prone to age-related structural decline such as brain atrophy. In this work, we present a new deep learning method called GRACE, which stands for General, Rapid, And Comprehensive whole-hEad tissue segmentation. GRACE is trained and validated on a novel dataset that consists of 177 manually corrected MR-derived reference segmentations that have undergone meticulous manual review. Each T1-weighted MRI volume is segmented into 11 tissue types, including white matter, grey matter, eyes, cerebrospinal fluid, air, blood vessel, cancellous bone, cortical bone, skin, fat, and muscle. To the best of our knowledge, this work contains the largest manually corrected dataset to date in terms of number of MRIs and segmented tissues. GRACE outperforms five freely available software tools and a traditional 3D U-Net on a five-tissue segmentation task. On this task, GRACE achieves an average Hausdorff Distance of 0.21, which exceeds the runner-up at an average Hausdorff Distance of 0.36. GRACE can segment a whole-head MRI in about 3 seconds, while the fastest software tool takes about 3 minutes. In summary, GRACE segments a spectrum of tissue types from older adults T1-MRI scans at favorable accuracy and speed. The trained GRACE model is optimized on older adult heads to enable high-precision modeling in age-related brain disorders. To support open science, the GRACE code and trained weights will be made available online and open to the research community upon publication.

Lossy compression introduces artifacts, and many conventional in-loop filters have been adopted in the AV1 standard to reduce these artifacts. Researchers have explored deep learning-based filters to remove artifacts in the compression loop. However, the high computational complexity of CNN-based filters remains a challenge. In this paper, a Texture- and Motion-Aware Perception (TMAP) in-loop filter is proposed to addresses this issue by selectively applying CNNs to texture-rich and high-motion regions, while utilizing non-learning methods to detect these regions. The proposed method introduces a new CNN structure, the Dense-Dual-Field Network (DDFN), which leverages a larger receptive field to enhance the quality of reconstructed frames by incorporating more contextual information. Furthermore, to improve perceptual quality, a novel loss function integrating wavelet-based perceptual information is presented. Experimental results demonstrate the superiority of our proposed models over other lightweight CNN models, and the effectiveness of the perceptual loss function is validated using the VMAF metric.


A handheld near-infrared optical scanner (NIROS) was recently developed to map for effective changes in oxy- and deoxyhemoglobin concentration in diabetic foot ulcers (DFUs) across weeks of treatment. Herein, a coregistration and image segmentation approach was implemented to overlay hemoglobin maps onto the white light images of ulcers. Validation studies demonstrated over 97% accuracy in coregistration. Coregistration was further applied to a healing DFU across weeks of healing. The potential to predict changes in wound healing was observed when comparing the coregistered and segmented hemoglobin concentration area maps to the visual area of the wound.

Advancements in brain imaging and stimulation have revolutionized our understanding of the human brain, its functioning, and the associated disorders. However, the historical male-dominated culture and practice in this field and the underrepresentation of women researchers and leaders could curb its development and limit its full potential. As such, this Research Topic, “Women in brain imaging and stimulation,” aims to showcase the work led by female researchers in the field and highlight their scholarship and scientific achievements at the frontier of interdisciplinary research of brain imaging and stimulation.

Neural network pruning is an essential technique for reducing the size and complexity of deep neural networks, enabling large-scale models on devices with limited resources. However, existing pruning approaches heavily rely on training data for guiding the pruning strategies, making them ineffective for federated learning over distributed and confidential datasets. Additionally, the memory- and computation-intensive pruning process becomes infeasible for recourse-constrained devices in federated learning. To address these challenges, we propose FedTiny, a distributed pruning framework for federated learning that generates specialized tiny models for memory- and computing-constrained devices. We introduce two key modules in FedTiny to adaptively search coarse- and finer-pruned specialized models to fit deployment scenarios with sparse and cheap local computation. First, an adaptive batch normalization selection module is designed to mitigate biases in pruning caused by the heterogeneity of local data. Second, a lightweight progressive pruning module aims to finer prune the models under strict memory and computational budgets, allowing the pruning policy for each layer to be gradually determined rather than evaluating the overall model structure. The experimental results demonstrate the effectiveness of FedTiny, which outperforms state-of-the-art approaches, particularly when compressing deep models to extremely sparse tiny models. FedTiny achieves an accuracy improvement of 2.61% while significantly reducing the computational cost by 95.91% and the memory footprint by 94.01% compared to state-of-the-art methods.

Model calibration measures the agreement between the predicted probability estimates and the true correctness likelihood. Proper model calibration is vital for high-risk applications. Unfortunately, modern deep neural networks are poorly calibrated, compromising trustworthiness and reliability. Medical image segmentation particularly suffers from this due to the natural uncertainty of tissue boundaries. This is exasperated by their loss functions, which favor overconfidence in the majority classes. We address these challenges with DOMINO, a domain-aware model calibration method that leverages the semantic confusability and hierarchical similarity between class labels. Our experiments demonstrate that our DOMINO-calibrated deep neural networks outperform non-calibrated models and state-of-the-art morphometric methods in head image segmentation. Our results show that our method can consistently achieve better calibration, higher accuracy, and faster inference times than these methods, especially on rarer classes. This performance is attributed to our domain-aware regularization to inform semantic model calibration. These findings show the importance of semantic ties between class labels in building confidence in deep learning models. The framework has the potential to improve the trustworthiness and reliability of generic medical image segmentation models.

Extracellular amyloid plaques in gray matter are the earliest pathological marker for Alzheimer’s disease (AD), followed by abnormal intraneuronal tau protein accumulation. Diffusion MRI (dMRI) is an imaging technique that estimates diffusion properties of the extracellular and tissue compartments. The link between diffusion changes and amyloid and tau pathology in gray matter is not well understood. We aimed to characterize the relationship between diffusion measures and amyloid and tau deposits in the gray matter across Braak stages, as assessed by tau PET imaging, in mild cognitive impairment (MCI) and AD patients.

Diabetic retinopathy (DR) is a leading cause of blindness in American adults. If detected, DR can be treated to prevent further damage causing blindness. There is an increasing interest in developing artificial intelligence (AI) technologies to help detect DR using electronic health records. The lesion-related information documented in fundus image reports is a valuable resource that could help diagnoses of DR in clinical decision support systems. However, most studies for AI-based DR diagnoses are mainly based on medical images; there is limited studies to explore the lesion-related information captured in the free text image reports.

A body of studies has proposed to obtain high-quality images from low-dose and noisy Computed Tomography (CT) scans for radiation reduction. However, these studies are designed for population-level data without considering the variation in CT devices and individuals, limiting the current approaches' performance, especially for ultra-low-dose CT imaging. Here, we proposed PIMA-CT, a physical anthropomorphic phantom model integrating an unsupervised learning framework, using a novel deep learning technique called Cyclic Simulation and Denoising (CSD), to address these limitations. We first acquired paired low-dose and standard-dose CT scans of the phantom and then developed two generative neural networks: noise simulator and denoiser. The simulator extracts real low-dose noise and tissue features from two separate image spaces (e.g., low-dose phantom model scans and standard-dose patient scans) into a unified feature space. Meanwhile, the denoiser provides feedback to the simulator on the quality of the generated noise. In this way, the simulator and denoiser cyclically interact to optimize network learning and ease the denoiser to simultaneously remove noise and restore tissue features. We thoroughly evaluate our method for removing both real low-dose noise and Gaussian simulated low-dose noise. The results show that CSD outperforms one of the state-of-the-art denoising algorithms without using any labeled data (actual patients' low-dose CT scans) nor simulated low-dose CT scans. This study may shed light on incorporating physical models in medical imaging, especially for ultra-low level dose CT scans restoration.








In addition to the well-established somatotopy in the pre- and post-central gyrus, there is now strong evidence that somatotopic organization is evident across other regions in the sensorimotor network. This raises several experimental questions: To what extent is activity in the sensorimotor network effector-dependent and effector-independent? How important is the sensorimotor cortex when predicting the motor effector? Is there redundancy in the distributed somatotopically organized network such that removing one region has little impact on classification accuracy? To answer these questions, we developed a novel experimental approach. fMRI data were collected while human subjects performed a precisely controlled force generation task separately with their hand, foot, and mouth. We used a simple linear iterative clustering (SLIC) algorithm to segment whole-brain beta coefficient maps to build an adaptive brain parcellation and then classified effectors using extreme gradient boosting (XGBoost) based on parcellations at various spatial resolutions. This allowed us to understand how data-driven adaptive brain parcellation granularity altered classification accuracy. Results revealed effector-dependent activity in regions of the post-central gyrus, precentral gyrus, and paracentral lobule. SMA, regions of the inferior and superior parietal lobule, and cerebellum each contained effector-dependent and effector-independent representations. Machine learning analyses showed that increasing the spatial resolution of the data-driven model increased classification accuracy, which reached 94% with over 1,755 supervoxels. Our SLIC-based supervoxel parcellation outperformed classification analyses using established brain templates and random simulations. Occlusion experiments further demonstrated redundancy across the sensorimotor network when classifying effectors. Our observations extend our understanding of effector-dependent and effector-independent organization within the human brain and provide new insight into the functional neuroanatomy required to predict the motor effector used in a motor control task.

Background and Objectives: Prediction of decline to dementia using objective biomarkers in high-risk patients with amnestic mild cognitive impairment (aMCI) has immense utility. Our objective was to use multimodal MRI to 1) determine whether accurate and precise prediction of dementia conversion could be achieved using baseline data alone, and 2) generate a map of the brain regions implicated in longitudinal decline to dementia.
Methods: Participants meeting criteria for aMCI at baseline (N=55) were classified at follow-up as remaining stable/improved in their diagnosis (N=41) or declined to dementia (N=14). Baseline T1 structural MRI and resting-state fMRI (rsfMRI) were combined and a semi-supervised support vector machine (SVM) which separated stable participants from those who decline at follow-up with maximal margin. Cross-validated model performance metrics and MRI feature weights were calculated to include the strength of each brain voxel in its ability to distinguish the two groups.
Results: Total model accuracy for predicting diagnostic change at follow-up was 92.7% using baseline T1 imaging alone, 83.5% using rsfMRI alone, and 94.5% when combining T1 and rsfMRI modalities. Feature weights that survived the p<0.01 threshold for separation of the two groups revealed the strongest margin in the combined structural and functional regions underlying the medial temporal lobes in the limbic system.
Discussion: An MRI-driven SVM model demonstrates accurate and precise prediction of later dementia conversion in aMCI patients. The multi-modal regions driving this prediction were the strongest in the medial temporal regions of the limbic system, consistent with literature on the progression of Alzheimer’s disease.

Recently, deep neural networks have demonstrated comparable and even better performance than board-certified ophthalmologists in well-annotated datasets. However, the diversity of retinal imaging devices poses a significant challenge: domain shift, which leads to performance degradation when applying the deep learning models trained on one domain to new testing domains. In this paper, we propose a multi-scale input along with multiple domain adaptors applied hierarchically in both feature and output spaces. The proposed training strategy and novel unsupervised domain adaptation framework, called Collaborative Adversarial Domain Adaptation (CADA), can effectively overcome the challenge. Multi-scale inputs can reduce the information loss due to the pooling layers used in the network for feature extraction, while our proposed CADA is an interactive paradigm that presents an exquisite collaborative adaptation through both adversarial learning and ensembling weights at different network layers. In particular, to produce a better prediction for the unlabeled target domain data, we simultaneously achieve domain invariance and model generalizability via adversarial learning at multi-scale outputs from different levels of network layers and maintaining an exponential moving average (EMA) of the historical weights during training. Without annotating any sample from the target domain, multiple adversarial losses in encoder and decoder layers guide the extraction of domain-invariant features to confuse the domain classifier. Meanwhile, the ensembling of weights via EMA reduces the uncertainty of adapting multiple discriminator learning. Comprehensive experimental results demonstrate that our CADA model incorporating multi-scale input training can overcome performance degradation and outperform state-of-the-art domain adaptation methods in segmenting retinal optic disc and cup from fundus images stemming from the REFUGE, Drishti-GS, and Rim-One-r3 datasets. The code will be available at https://github.com/cswin/CADA

This article deals with approximating steady-state particle-resolved fluid flow around a fixed particle of interest under the influence of randomly distributed stationary particles in a dispersed multiphase setup using convolutional neural network (CNN). The considered problem involves rotational symmetry about the mean velocity (streamwise) direction. Thus, this work enforces this symmetry using SE(3)-equivariant, special Euclidean group of dimension 3, CNN architecture, which is translation and three-dimensional rotation equivariant. This study mainly explores the generalization capabilities and benefits of a SE(3)-equivariant network. Accurate synthetic flow fields for Reynolds number and particle volume fraction combinations spanning over a range of [86.22, 172.96] and [0.11, 0.45], respectively, are produced with careful application of symmetry-aware data-driven approach.

Fluid flow around a random distribution of stationary spherical particles is a problem of substantial importance in the study of dispersed multiphase flows. In this paper we present a machine learning methodology using Generative Adversarial Network framework and Convolutional Neural Network architecture to recreate particle-resolved fluid flow around a random distribution of monodispersed particles. The model was applied to various Reynolds number and par- ticle volume fraction combinations spanning over a range of [2.69, 172.96] and [0.11, 0.45] respectively. Test performance of the model for the studied cases is very promising.
Keywords: Pseudo-turbulence, Multiphase Flow prediction, Generative Adversarial Network (GAN), Convolutional Neural Network (CNN)


Alzheimer's disease is the leading cause of dementia. The long progression period in Alzheimer's disease provides a possibility for patients to get early treatment by having routine screenings. However, current clinical diagnostic imaging tools do not meet the specific requirements for screening procedures due to high cost and limited availability. In this work, we took the initiative to evaluate the retina, especially the retinal vasculature, as an alternative for conducting screenings for dementia patients caused by Alzheimer's disease. Highly modular machine learning techniques were employed throughout the whole pipeline. Utilizing data from the UK Biobank, the pipeline achieved an average classification accuracy of 82.44%. Besides the high classification accuracy, we also added a saliency analysis to strengthen this pipeline's interpretability. The saliency analysis indicated that within retinal images, small vessels carry more information for diagnosing Alzheimer's diseases, which aligns with related studies.




Transcranial direct current stimulation (tDCS) is widely investigated as a therapeutic tool to enhance cognitive function in older adults with and without neurodegenerative disease. Prior research demonstrates that electric current delivery to the brain can vary significantly across individuals. Quantification of this variability could enable person-specific optimization of tDCS outcomes. This pilot study used machine learning and MRI-derived electric field models to predict working memory improvements as a proof of concept for precision cognitive intervention.
Fourteen healthy older adults received 20 minutes of 2 mA tDCS stimulation (F3/F4) during a two-week cognitive training intervention. Participants performed an N-back working memory task pre-/post-intervention. MRI-derived current models were passed through a linear Support Vector Machine (SVM) learning algorithm to characterize crucial tDCS current components (intensity and direction) that induced working memory improvements in tDCS responders versus non-responders.
SVM models of tDCS current components had 86% overall accuracy in classifying treatment responders vs. non-responders, with current intensity producing the best overall model differentiating changes in working memory performance. Median current intensity and direction in brain regions near the electrodes were positively related to intervention responses.
This study provides the first evidence that pattern recognition analyses of MRI-derived tDCS current models can provide individual prognostic classification of tDCS treatment response with 86% accuracy. Individual differences in current intensity and direction play important roles in determining treatment response to tDCS. These findings provide important insights into mechanisms of tDCS response as well as proof of concept for future precision dosing models of tDCS intervention.



A handheld near-infrared optical scanner (NIROS) was recently developed to map for effective changes in oxy- and deoxyhemoglobin concentration in diabetic foot ulcers (DFUs) across weeks of treatment. Herein, a coregistration and image segmentation approach was implemented to overlay hemoglobin maps onto the white light images of ulcers. Validation studies demonstrated over 97% accuracy in coregistration. Coregistration was further applied to a healing DFU across weeks of healing. The potential to predict changes in wound healing was observed when comparing the coregistered and segmented hemoglobin concentration area maps to the visual area of the wound.

OBJECTIVES/GOALS: Spinal cord stimulation (SCS) is an intervention for patients with chronic back pain. Technological advances have led to renewed optimism in the field, but mechanisms of action in the brain remain poorly understood. We hypothesize that SCS outcomes will be associated with changes in neural oscillations. METHODS/STUDY POPULATION: The goal of our team project is to test patients who receive SCS at 3 times points: baseline, at day 7 during the trial period, and day 180 after a permanent system has been implanted. At each time point participants will complete 10 minutes of eyes closed, resting electroencephalography (EEG). EEG will be collected with the ActiveTwo system, a 128-electrode cap, and a 256 channel AD box from BioSemi. Traditional machine learning methods such as support vector machine and more complex models including deep learning will be used to generate interpretable features within resting EEG signals. RESULTS/ANTICIPATED RESULTS: Through machine learning, we anticipate that SCS will have a significant effect on resting alpha and beta power in sensorimotor cortex. DISCUSSION/SIGNIFICANCE OF IMPACT: This collaborative project will further the application of machine learning in cognitive neuroscience and allow us to better understand how therapies for chronic pain alter resting brain activity.

Diabetic Retinopathy (DR) represents a highly-prevalent complication of diabetes in which individuals suffer from damage to the blood vessels in the retina. The disease manifests itself through lesion presence, starting with microaneurysms, at the nonproliferative stage before being characterized by neovascularization in the proliferative stage. Retinal specialists strive to detect DR early so that the disease can be treated before substantial, irreversible vision loss occurs. The level of DR severity indicates the extent of treatment necessary - vision loss may be preventable by effective diabetes management in mild (early) stages, rather than subjecting the patient to invasive laser surgery. Using artificial intelligence (AI), highly accurate and efficient systems can be developed to help assist medical professionals in screening and diagnosing DR earlier and without the full resources that are available in specialty clinics. In particular, deep learning facilitates diagnosis earlier and with higher sensitivity and specificity. Such systems make decisions based on minimally handcrafted features and pave the way for personalized therapies. Thus, this survey provides a comprehensive description of the current technology used in each step of DR diagnosis. First, it begins with an introduction to the disease and the current technologies and resources available in this space. It proceeds to discuss the frameworks that different teams have used to detect and classify DR. Ultimately, we conclude that deep learning systems offer revolutionary potential to DR identification and prevention of vision loss.

Objective and quantitative assessment of fundus image quality is essential for the diagnosis of retinal diseases. The major factors in fundus image quality assessment are image artifact, clarity, and field definition. Unfortunately, most of existing quality assessment methods focus on the quality of overall image, without interpretable quality feedback for real-time adjustment. Furthermore, these models are often sensitive to the specific imaging devices, and cannot generalize well under different imaging conditions. This paper presents a new multi-task domain adaptation framework to automatically assess fundus image quality. The proposed framework provides interpretable quality assessment with both quantitative scores and quality visualization for potential real-time image recapture with proper adjustment. In particular, the present approach can detect optic disc and fovea structures as landmarks, to assist the assessment through coarse-to-fine feature encoding. The framework also exploit semi-tied adversarial discriminative domain adaptation to make the model generalizable across different data sources. Experimental results demonstrated that the proposed algorithm outperforms different state-of-the-art approaches and achieves an area under the ROC curve of 0.9455 for the overall quality classification.



This paper presents a deep neural network (DNN) based approach for predicting mean and peak wind pressure coefficients on the surface of a scale model low-rise, gable roof residential building. Pressure data were collected on the model at multiple prescribed wind directions and terrain roughnesses. The resultant pressure coefficients quantified from a subset of these directions and terrains were used to train an DNN to predict coefficients for directions and terrains excluded from the training. The approach is able to leverage a variety of input conditions to predict pressure coefficients with high accuracy, while the prior work has limited flexibility with the number of input variables and yielded lower prediction accuracy. A two-step nested DNN procedure is introduced to improve the prediction of peak coefficients. The optimal correlation coefficients of return predictions were 0.9993 and 0.9964, for mean and peak coefficient prediction, respectively. The concept of super resolution based on global prediction was also discussed. With a sufficiently large database, the proposed DNN-based approach can augment existing experimental methods to improve the yield of knowledge while reducing the number of tests required to gain that knowledge.

Diabetic Retinopathy (DR) is the most common cause of avoidable vision loss, predominantly affecting the working-age population across the globe. Screening for DR, coupled with timely consultation and treatment, is a globally trusted policy to avoid vision loss. However, implementation of DR screening programs is challenging due to the scarcity of medical professionals able to screen a growing global diabetic population at risk for DR. Computer-aided disease diagnosis in retinal image analysis could provide a sustainable approach for such large-scale screening effort. The recent scientific advances in computing capacity and machine learning approaches provide an avenue for biomedical scientists to reach this goal. Aiming to advance the state-of-the-art in automatic DR diagnosis, a grand challenge on “Diabetic Retinopathy – Segmentation and Grading” was organized in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI - 2018). In this paper, we report the set-up and results of this challenge that is primarily based on Indian Diabetic Retinopathy Image Dataset (IDRiD). There were three principal sub-challenges: lesion segmentation, disease severity grading, and localization of retinal landmarks and segmentation. These multiple tasks in this challenge allow to test the generalizability of algorithms, and this is what makes it different from existing ones. It received a positive response from the scientific community with 148 submissions from 495 registrations effectively entered in this challenge. This paper outlines the challenge, its organization, the dataset used, evaluation methods and results of top-performing participating solutions. The top-performing approaches utilized a blend of clinical information, data augmentation, and an ensemble of models. These findings have the potential to enable new developments in retinal image analysis and image-based DR screening in particular.

Glaucoma is one of the leading causes of irreversible but preventable blindness in working age populations. Color fundus photography (CFP) is the most cost-effective imaging modality to screen for retinal disorders. However, its application to glaucoma has been limited to the computation of a few related biomarkers such as the vertical cup-to-disc ratio. Deep learning approaches, although widely applied for medical image analysis, have not been extensively used for glaucoma assessment due to the limited size of the available data sets. Furthermore, the lack of a standardize benchmark strategy makes difficult to compare existing methods in a uniform way. In order to overcome these issues we set up the Retinal Fundus Glaucoma Challenge, REFUGE (https://refuge.grand-challenge.org), held in conjunction with MICCAI 2018. The challenge consisted of two primary tasks, namely optic disc/cup segmentation and glaucoma classification. As part of REFUGE, we have publicly released a data set of 1200 fundus images with ground truth segmentations and clinical glaucoma labels, currently the largest existing one. We have also built an evaluation framework to ease and ensure fairness in the comparison of different models, encouraging the development of novel techniques in the field. 12 teams qualified and participated in the online challenge. This paper summarizes their methods and analyzes their corresponding results. In particular, we observed that two of the top-ranked teams outperformed two human experts in the glaucoma classification task. Furthermore, the segmentation results were in general consistent with the ground truth annotations, with complementary outcomes that can be further exploited by ensembling the results.





Deep convolutional neural networks offer state-of-the-art performance for medical image analysis. However, their architectures are manually designed for particular problems. On the one hand, a manual designing process requires many trials to tune a large number of hyperparameters and is thus quite a time-consuming task. On the other hand, the fittest hyperparameters that can adapt to source data properties (e.g., sparsity, noisy features) are not able to be quickly identified for target data properties. For instance, the realistic noise in medical images is usually mixed and complicated, and sometimes unknown, leading to challenges in applying existing methods directly and creating effective denoising neural networks easily. In this paper, we present a Genetic Algorithm (GA)-based network evolution approach to search for the fittest genes to optimize network structures automatically. We expedite the evolutionary process through an experience-based greedy exploration strategy and transfer learning. Our evolutionary algorithm procedure has flexibility, which allows taking advantage of current state-of-the-art modules (e.g., residual blocks) to search for promising neural networks. We evaluate our framework on a classic medical image analysis task: denoising. The experimental results on computed tomography perfusion (CTP) image denoising demonstrate the capability of the method to select the fittest genes for building high-performance networks, named EvoNets. Our results outperform state-of-the-art methods consistently at various noise levels.

Computed Tomography Perfusion (CTP) imaging is a cost-effective and fast approach to provide diagnostic images for acute stroke treatment. Its cine scanning mode allows the visualization of anatomic brain structures and blood flow; however, it requires contrast agent injection and continuous CT scanning over an extended time. In fact, the accumulative radiation dose to patients will increase health risks such as skin irritation, hair loss, cataract formation, and even cancer. Solutions for reducing radiation exposure include reducing the tube current and/or shortening the X-ray radiation exposure time. However, images scanned at lower tube currents are usually accompanied by higher levels of noise and artifacts. On the other hand, shorter X-ray radiation exposure time with longer scanning intervals will lead to image information that is insufficient to capture the blood flow dynamics between frames. Thus, it is critical for us to seek a solution that can preserve the image quality when the tube current and the temporal frequency are both low. We propose STIR-Net in this paper, an end-to-end spatial-temporal convolutional neural network structure, which exploits multi-directional automatic feature extraction and image reconstruction schema to recover high-quality CT slices effectively. With the inputs of low-dose and low-resolution patches at different cross-sections of the spatio-temporal data, STIR-Net blends the features from both spatial and temporal domains to reconstruct high-quality CT volumes. In this study, we finalize extensive experiments to appraise the image restoration performance at different levels of tube current and spatial and temporal resolution scales.The results demonstrate the capability of our STIR-Net to restore high-quality scans at as low as 11% of absorbed radiation dose of the current imaging protocol, yielding an average of 10% improvement for perfusion maps compared to the patch-based log likelihood method.

The choroid layer is a vascular layer in human retina and its main function is to provide oxygen and support to the retina. Various studies have shown that the thickness of the choroid layer is correlated with the diagnosis of several ophthalmic diseases. For example, diabetic macular edema (DME) is a leading cause of vision loss in patients with diabetes. Despite contemporary advances, automatic segmentation of the choroid layer remains a challenging task due to low contrast, inhomogeneous intensity, inconsistent texture and ambiguous boundaries between the choroid and sclera in Optical Coherence Tomography (OCT) images. The majority of currently implemented methods manually or semi-automatically segment out the region of interest. While many fully automatic methods exist in the context of choroid layer segmentation, more effective and accurate automatic methods are required in order to employ these methods in the clinical sector. This paper proposed and implemented an automatic method for choroid layer segmentation in OCT images using deep learning and a series of morphological operations. The aim of this research was to segment out Bruch’s Membrane (BM) and choroid layer to calculate the thickness map. BM was segmented using a series of morphological operations, whereas the choroid layer was segmented using a deep learning approach as more image statistics were required to segment accurately. Several evaluation metrics were used to test and compare the proposed method against other existing methodologies. Experimental results showed that the proposed method greatly reduced the error rate when compared with the other state-of-the-art methods.








The advent of automatic tracing and reconstruction technology has led to a surge in the number of neurons 3D reconstruction data and consequently the neuromorphology research. However, the lack of machine-driven annotation schema to automatically detect the types of the neurons based on their morphology still hinders the development of this branch of science. Neuromorphology is important because of the interplay between the shape and functionality of neurons and the far-reaching impact on the diagnostics and therapeutics in neurological disorders. This survey paper provides a comprehensive research in the field of automatic neurons classification and presents the existing challenges, methods, tools, and future directions for automatic neuromorphology analytics. We summarize the major automatic techniques applicable in the field and propose a systematic data processing pipeline for automatic neuron classification, covering data capturing, preprocessing, analyzing, classification, and retrieval. Various techniques and algorithms in machine learning are illustrated and compared to the same dataset to facilitate ongoing research in the field.

The retinal vessel is one of the determining factors in an ophthalmic examination. Automatic extraction of retinal vessels from low-quality retinal images still remains a challenging problem. In this paper, we propose a robust and effective approach that qualitatively improves the detection of low-contrast and narrow vessels. Rather than using the pixel grid, we use a superpixel as the elementary unit of our vessel segmentation scheme. We regularize this scheme by combining the geometrical structure, texture, color, and space information in the superpixel graph. And the segmentation results are then refined by employing the efficient minimum spanning superpixel tree to detect and capture both global and local structure of the retinal images. Such an effective and structure-aware tree detector significantly improves the detection around the pathologic area. Experimental results have shown that the proposed technique achieves advantageous connectivity-area-length (CAL) scores of 80.92% and 69.06% on two public datasets, namely, DRIVE and STARE, thereby outperforming state-of-the-art segmentation methods. In addition, the tests on the challenging retinal image database have further demonstrated the effectiveness of our method. Our approach achieves satisfactory segmentation performance in comparison with state-of-the-art methods. Our technique provides an automated method for effectively extracting the vessel from fundus images

Convolutional neural networks offer state-of-the-art performance for medical image denoising. However, their architectures are manually designed for different noise types. The realistic noise in medical images is usually mixed and complicated, and sometimes unknown, leading to challenges in creating effective denoising neural networks. In this paper, we present a Genetic Algorithm (GA)-based network evolution approach to search for the fittest genes to optimize network structures. We expedite the evolutionary process through an experience-based greedy exploration strategy and transfer learning. The experimental results on computed tomography perfusion (CTP) images denoising demonstrate the capability of the method to select the fittest genes for building high-performance networks, named EvoNets, and our results compare favorably with state-of-the-art methods

The quality of fundus images is critical for diabetic retinopathy diagnosis. The evaluation of fundus image quality can be affected by several factors, including image artifact, clarity, and field definition. In this paper, we propose a multi-task deep learning framework for automated assessment of fundus image quality. The network can classify whether an image is gradable, together with interpretable information about quality factors. The proposed method uses images in both rectangular and polar coordinates, and fine-tunes the network from trained model grading of diabetic retinopathy. The detection of optic disk and fovea assists learning the field definition task through coarse-to-fine feature encoding. The experimental results demonstrate that our framework outperform single-task convolutional neural networks and reject ungradable images in automated diabetic retinopathy diagnostic systems.










Optical coherence tomography (OCT) is an immersive technique for depth analysis of retinal layers. Automatic choroid layer segmentation is a challenging task because of the low contrast inputs. Existing methodologies carried choroid layer segmentation manually or semi-automatically. In this paper, we proposed automated choroid layer segmentation based on normalized cut algorithm, which aims at extracting the global impression of images and treats the segmentation as a graph partitioning problem. Due to the complexity of the layering structure of retinal layers and choroid layer, we employed a series of preprocessing to make the cut more deterministic and accurate. The proposed method divided the image into several patches and ran the normalized cut on every image patch separately. The aim was to avoid insignificant vertical cuts and focus on horizontal cutting. After processing every patch, we acquired a global cut on the original image by combining all the patches. Later we measured the choroidal thickness which is highly helpful in the diagnosis of several retinal diseases. The results were computed on a total of 525 images of 21 real patients. Experimental results showed that the mean relative error rate of the proposed method was around 0.4 as the compared the manual segmentation performed by the experts.

Computed tomography perfusion (CTP) is one of the most widely used imaging modality for cerebrovascular disease diagnosis and treatment, especially in emergency situations. While cerebral CTP is ca- pable of quantifying the blood flow dynamics by continuous scanning at a focused region of the brain, the associated excessive radiation increases the patients' risk levels of developing cancer. To reduce the necessary radiation dose in CTP, decreasing the temporal sampling frequency is one promising direction. In this paper, we propose STAR, an end-to- end Spatio-Temporal Architecture for super-Resolution to significantly reduce the necessary scanning time and therefore radiation exposure. The inputs into STAR are multi-directional 2D low-resolution spatio- temporal patches at different cross sections over space and time. Via training multiple direction networks followed by a conjoint reconstruc- tion network, our approach can produce high-resolution spatio-temporal volumes. The experiment results demonstrate the capability of STAR to maintain the image quality and accuracy of cerebral hemodynamic parameters at only one-third of the original scanning time.




Cardiovascular disease is one of the leading causes of death in the United States. It is critical to identify the risk factors associated with cardiovascular diseases and to alert individuals before they experience a heart attack. In this paper we propose RFMiner, a risk factor discovery and mining framework for identifying significant risk factors using integrated measures. We provide the blueprints for accurately predicting the possibility of heart attacks in the near future while identifying notable risk factors - especially the factors which are not well recognized.

Morphological retrieval is an effective approach to explore large-scale neuronal databases, as the morphology is correlated with neuronal types, regions, functions, etc. In this paper, we focus on the neuron identification and analysis via morphological retrieval. In our proposed framework, multiple features are extracted to represent 3D neuron data. Because each feature reflects different levels of similarity between neurons, we group features into different hierarchies to compute the similarity matrix. Then, compact binary codes are generated from hierarchical features for efficient similarity search. Since neuronal cells usually have tree-topology structure, it is hard to distinguish different types of neurons simply via traditional binary coding or hashing methods based on Euclidean distance metric and/or linear hyperplanes. Therefore, we employ an asymmetric binary coding strategy based on the maximum inner product search (MIPS), which not only makes it easier to learn the binary coding functions, but also preserves the non-linear characteristics of the neuron morphological data. We evaluate the proposed method on more than 17,000 neurons, by validating the retrieved neurons with associated cell types and brain regions. Experimental results show the superiority of our approach in neuron morphological retrieval compared with other state-of-the-art methods. Moreover, we demonstrate its potential use cases in the identification and analysis of neuron characteristics from large neuron databases.

Segmentation of abdominal adipose tissues (AAT) into subcutaneous adipose tissues (SAT) and visceral adipose tissues (VAT) is of crucial interest for managing the obesity. Previous methods with raw or hand-crafted features rarely work well on large-scale subject cohorts, because of the inhomogeneous image intensities, artifacts and the diverse distributions of VAT. In this paper, we propose a novel two-stage coarse-to-fine algorithm for AAT seg- mentation. In the first stage, we formulate the AAT segmentation task as a pixel-wise classification problem. First, three types of features, intensity, spatial and contextual fea- tures, are adopted. Second, a new type of deep neural network, named multi-scale deep neural network (MSDNN), is provided to extract high-level features. In the second stage, to improve the segmentation accuracy, we refine coarse segmentation results by determining the internal boundary of SAT based on coarse segmentation results and continuous of SAT internal boundary. Finally, we demonstrate the efficacy of our algorithm for both 2D and 3D cases on a wide population range. Compared with other algorithms, our method is not only more suitable for large-scale dataset, but also achieves better segmentation results. Fur- thermore, our system takes about 2 seconds to segment an abdominal image, which implies potential clinical applications.


Stroke is the leading cause of long-term disability and the second leading cause of mortality in the world, and exerts an enormous burden on the public health. CT remains one of the most widely used imaging modality for stroke diagnosis. However when coupled with CT perfusion, the excessive radiation exposure in repetitive imaging to assess treatment response and prognosis has raised significant public concerns regarding its potential hazards to both short- and longterm health outcomes. Tensor total variation has been proposed to reduce the necessary radiation dose in CT perfusion without comprising the image quality by fusing the information of the local anatomical structure with the temporal blood flow model. However the local search in the framework fails to leverage the non-local information in the spatio-temporal data. In this paper, we propose TENDER, an efficient framework of non-local tensor deconvolution to maintain the accuracy of the hemodynamic parameters and the diagnostic reliability in low radiation dose CT perfusion. The tensor total variation is extended using non-local spatio-temporal cubics for regularization to integrate contextual and non-local information. We also propose an efficient framework consisting of fast nearest neighbor search, accelerated optimization and parallel computing to improve the efficiency and scalability of the non-local spatio-temporal algorithm. Evaluations on clinical data of subjects with cerebrovascular disease and normal subjects demonstrate the advantage of non-local tensor deconvolution for reducing radiation dose in CT perfusion.

The explosive growth and widespread accessibility of digital health data have led to a surge of research activity in the healthcare and data sciences fields. The conventional approaches for health data management have achieved limited success as they are incapable of handling the huge amount of complex data with high volume, high velocity, and high variety. This article presents a comprehensive overview of the existing challenges, techniques, and future directions for computational health informatics in the big data age, with a structured analysis of the historical and state-of-the-art methods. We have summarized the challenges into four Vs (i.e., volume, velocity, variety, and veracity) and proposed a systematic data-processing pipeline for generic big data in health informatics, covering data capturing, storing, sharing, analyzing, searching, and decision support. Specifically, numerous techniques and algorithms in machine learning are categorized and compared. On the basis of this material, we identify and discuss the essential prospects lying ahead for computational health informatics in this big data age.

Clinicians employ visual inspection of the wound and reduction in its size over time to monitor its healing process. Although these are standard clinical assessments, there is a need to develop a physiological approach that can map sub-surface tissue oxygenation at and around the wound region. Recently, a non-contact, portable, hand-held near infrared optical scanner (NIROS) has been developed to functionally image wound sites and differentiate healing from non-healing in lower extremity ulcers. Past studies using NIROS focused on differentiating healing from non-healing wounds based on NIR optical contrast between the wound and healthy surrounding tissue. However, these studies did not showcase the physiological changes in tissue oxygenation. Herein, NIROS has been modified to perform multi wavelength imaging in order to obtain the oxy and deoxy- hemoglobin maps of the wound and its surroundings. Clinical studies are currently performed at two clinical sites in Miami on lower extremity ulcers (2, diabetic foot ulcers (DFUs) and 4 venous leg ulcers (VLUs to date). Preliminary results have shown changes in oxy- and deoxy-hemoglobin maps of the wound and background across weeks of the treatment process. Image segmentation studies quantified regions of varied tissue oxygenation around and beneath the wound to potentially determine sub-surface healing regions. Ongoing efforts involve systematic 8-week imaging studies to obtain physiological indicators of healing from hemodynamic studies of DFUs and VLUs.



Morphological retrieval is an effective approach to explore neurons' databases, as the morphology is correlated with neuronal types, regions, functions, etc. In this paper, we focus on the neuron identification and analysis via morphological retrieval. In our proposed framework, both global and local features are extracted to represent 3D neuron data. Then, compacted binary codes are generated from original features for efficient similarity search. As neuron cells usually have tree-topology structure, it is hard to distinguish different types of neuron simply via traditional binary coding or hashing methods based on Euclidean distance metric and/or linear hyperplanes. Thus, we propose a novel binary coding method based on the maximum inner product search (MIPS), which is not only more easier to learn the binary coding function, but also preserves the non-linear characteristics of neuron morphology data. We evaluate the proposed method on more than 17,000 neurons, by validating the retrieved neurons with associated cell types and brain regions. Experimental results show the superiority of our approach in neuron morphological retrieval compared with other state-of-the-art methods. Moreover, we demonstrate its potential use case in the identification and analysis of neuron characteristics.

With the goal of achieving low radiation exposure from medical imaging, computed tomography perfusion (CTP) introduces challenging problems for both image reconstruction and perfusion parameter estimation in the qualitative and quantitative analyses. Conventional approaches address the reconstruction and the estimation processes separately. Since the hemodynamic parameter maps have much lower dimensionality than the original sinogram data, estimating hemodynamic parameters directly from sinogram will further reduce radiation exposure and save computational resources to reconstruct the intermediate time-series images. In this work, we propose the first direct estimation framework for CTP that integrates the time-series image reconstruction, contrast conversion, hematocrit correction and hemodynamic parameter estimation in one optimization function, which is solved using an efficient algorithm. Evaluations on the digital brain perfusion phantom and a clinical acute stroke subject demonstrate that the proposed direct estimation framework boosts the estimation accuracy remarkably in CTP scanning with lower radiation exposure.


Near-Infrared (NIR) optical imaging can reveal tissue oxygenation of the wound, complementing the visual inspection of the surface granulation. Herein, graph cuts algorithm is applied to segment NIR images of the wound from its peripheries.

Lower extremity ulcers are one of the most common complications that not only affect many people around the world but also have huge impact on economy since a large amount of resources are spent for treatment and prevention of the diseases. Clinical studies have shown that reduction in the wound size of 40% within 4 weeks is an acceptable progress in the healing process. Quantification of the wound size plays a crucial role in assessing the extent of healing and determining the treatment process. To date, wound healing is visually inspected and the wound size is measured from surface images. The extent of wound healing internally may vary from the surface. A near-infrared (NIR) optical imaging approach has been developed for non-contact imaging of wounds internally and differentiating healing from non-healing wounds. Herein, quantitative wound size measurements from NIR and white light images are estimated using a graph cuts and region growing image segmentation algorithms. The extent of the wound healing from NIR imaging of lower extremity ulcers in diabetic subjects are quantified and compared across NIR and white light images. NIR imaging and wound size measurements can play a significant role in potentially predicting the extent of internal healing, thus allowing better treatment plans when implemented for periodic imaging in future.

Tensor total variation deconvolution has been recently proposed as a robust framework to accurately estimate the hemodynamic parameters in low-dose CT perfusion by fusing the local anatomicalstructure correlation and temporal blood flow continuation. However the locality property in the current framework constrains the search for anatomical structure similarities to the local neighborhood, missing the global and long-range correlations in the whole anatomical structure. This limitation has led to noticeable absence or artifact of delicate structures, including the critical indicators for the clinical diagnosis of cerebrovascular diseases. In this paper, we propose an extension of the TTV framework by introducing 4D non-local tensor total variation into the deconvolution to bridge the gap between non-adjacent regions of the same tissue classes. The non-local regularization using tensor total variation term is imposed on the spatio-temporal flow-scaled residue functions. An efficient algorithm and implementation of the non-local tensor total variation (NL-TTV) reduces the time complexity with fast similarity computation, accelerated optimization and parallel operations. Extensive evaluations on the clinical data with cerebrovascular diseases and normal subjects demonstrate the importance of non-local linkage and long-range connections for low-dose CT perfusion deconvolution.

Acute brain diseases such as acute strokes and transit ischemic attacks are the leading causes of mortality and morbidity worldwide, responsible for 9% of total death every year. ‘Time is brain' is a widely accepted concept in acute cerebrovascular disease treatment. Efficient and accurate computational framework for hemodynamic parameters estimation can save critical time for thrombolytic therapy. Meanwhile the high level of accumulated radiation dosage due to continuous image acquisition in CT perfusion (CTP) raised concerns on patient safety and public health. However, low-radiation leads to increased noise and artifacts which require more sophisticated and time-consuming algorithms for robust estimation. In this paper, we focus on developing a robust and efficient framework to accurately estimate the perfusion parameters at low radiation dosage. Specifically, we present a tensor total-variation (TTV) technique which fuses the spatial correlation of the vascular structure and the temporal continuation of the blood signal flow. An efficient algorithm is proposed to find the solution with fast convergence and reduced computational complexity. Extensive evaluations are carried out in terms of sensitivity to noise levels, estimation accuracy, contrast preservation, and performed on digital perfusion phantom estimation, as well as in-vivo clinical subjects. Our framework reduces the necessary radiation dose to only 8% of the original level and outperforms the state-of-art algorithms with peak signal-to-noise ratio improved by 32%. It reduces the oscillation in the residue functions, corrects overestimation of cerebral blood flow (CBF) and under-estimation of mean transit time (MTT), and maintains the distinction between the deficit and normal regions.

With the advent of the age for big data and complex structure, sparsity has been an important modeling tool in compressed sensing, machine learning, image processing, neuroscience and statistics. In the medical imaging field, sparsity methods have been successfully used in image reconstruction, image enhancement, image segmentation, anomaly detection, disease classification, and image database retrieval. Developing more powerful sparsity models for a large range of medical imaging and medical image analysis problems as well as efficient optimization and learning algorithm will keep being a main research topic in this field. The goal of this special issue is to publish original and high quality papers on innovation research and development in medical imaging and medical image analysis using sparsity techniques. This special issue will help advance the scientific research within the field of sparsity methods for medical imaging.

Enhancing perfusion maps in low-dose computed tomography perfusion (CTP) for cerebrovascular disease diagnosis is a challenging task, especially for lowcontrast tissue categories where infarct core and ischemic penumbra usually occur. Sparse perfusion deconvolution has been recently proposed to effectively improve the image quality and diagnostic accuracy of low-dose perfusion CT by extracting the complementary information from the high-dose perfusion maps to restore the low-dose using a joint spatio-temporal model. However the lowcontrast tissue classes where infarct core and ischemic penumbra are likely to occur in cerebral perfusion CT tend to be over-smoothed, leading to loss of essential biomarkers. In this paper, we propose a tissue-specific sparse deconvolution approach to preserve the subtle perfusion information in the low-contrast tissue classes. We first build tissue-specific dictionaries from segmentations of high-dose perfusion maps using online dictionary learning, and then perform deconvolution-based hemodynamic parameters estimation for block-wise tissue segments on the low-dose CTP data. Extensive validation on clinical datasets of patients with cerebrovascular disease demonstrates the superior performance of our proposed method compared to state-of-art, and potentially improve diagnostic accuracy by increasing the differentiation between normal and ischemic tissues in the brain




Stroke and cerebrovascular diseases are the leading cause of serious, long-term disability in the United States. Computed tomography perfusion (CTP) is one of the most widely accepted imaging modality for stroke care. However, the high radiation exposure of CTP has lead to increased cancer risk. Tensor total variation (TTV)[1] has been proposed to stabilize the quantification of perfusion parameters by integrating the anatomical structure correlation. Yet the locality limitation of the neighborhood region has led to noticeable absence or inflation of the delicate structures which are critical indicators for the clinical diagnosis. In this work, we propose a non-local tensor total variation (NL-TTV) deconvolution method to by incorporating the long-range dependency and the global connections in the spatio-temporal domain
4-D dynamic susceptibility contrast (DSC) magnetic resonance imaging (MRI) is a well-established perfusion technique for non-invasive characterization of tissue dynamics, with promising applications in assessing a wide range of diseases, as well as monitoring response of therapeutic interventions). DSC-MRI provides critical real-time information by tracking the first-pass of an injected contrast-agent (e.g. gadolinium) with T2*-weighted MRI. The spatio-temproal data, consisting of contrast concentration signals for each voxel of a volume, are deconvolved from the arterial input function (AIF) and then post-processed to generate perfusion parameter maps, typically including the cerebral blood flow (CBF), cerebral blood volume (CBV), mean transit time (MTT) and time to peak (TTP). The most popular deconvolution method is truncated singular value decomposition (TSVD)1,2 and its variants3 , which fail to exploit the spatio-temporal nature of the 4D data with both the anatomical structure and the temporal continuation. This work adapts and demonstrates the feasibility of a 4-D tensor total variation (TTV) deconvolution approach, which has been proposed for CT perfusion4 , to brain MR perfusion, with evaluation on synthetic data and clinical DSC-MRI data for glioblastomas, the most common type of brain cancer. The method is guaranteed to convergence to global optimal because of the convex cost function and presents a more elegant framework of total variation for the deconvolution, compared to recent efforts5,6 which either do not have a global optimal solution for the non-convex case or need to handcraft spatial and temporal regularization terms.


Arterial spin labeling MRI (ASL-MRI) can provide quantitative signals correlated to the cerebral blood flow and neural activity. However, the low signal-to-noise ratio in ASL requires repeated acquisitions to improve the signal reliability, leading to prolonged scanning time. At fewer repetitions, noise and corruptions arise due to motion and physiological artifacts, introducing errors into the cerebral blood flow estimation. We propose to recover the ASL-MRI data from the noisy and corrupted observations at shorter scanning time with a spatio-temporal low-rank total variation method. The low-rank approximation uses the similarity of the repetitive scans, and the total variation regularization considers the local spatial consistency. We compare with the state-of-art robust M-estimator for ASL cerebral blood flow map estimation. Validation on simulated and real data demonstrate the robustness of the proposed method at fewer scanning repetitions and with random corruption

Blood-brain barrier permeability (BBBP) measurements extracted from the perfusion computed tomography (PCT) using the Patlak model can be a valuable indicator to predict hemorrhagic transformation in patients with acute stroke. Unfortunately, the standard Patlak model based PCT requires excessive radiation exposure, which raised attention on radiation safety. Minimizing radiation dose is of high value in clinical practice but can degrade the image quality due to the introduced severe noise. The purpose of this work is to construct high quality BBBP maps from low-dose PCT data by using the brain structural similarity between different individuals and the relations between the high- and low-dose maps. The proposed sparse high-dose induced (shd-Patlak) model performs by building a high-dose induced prior for the Patlak model with a set of location adaptive dictionaries, followed by an optimized estimation of BBBP map with the prior regularized Patlak model. Evaluation with the simulated low-dose clinical brain PCT datasets clearly demonstrate that the shd-Patlak model can achieve more significant gains than the standard Patlak model with improved visual quality, higher fidelity to the gold standard and more accurate details for clinical analysis.

Acute brain diseases such as acute stroke and transit ischemic attacks are the leading causes of mortality and morbidity worldwide, responsible for 9% of total death every year. ‘Time is brain' is a widely accepted concept in acute cerebrovascular disease treatment. Efficient and accurate computational framework for hemodynamic parameters estimation can save critical time for thrombolytic therapy. Meanwhile the high level of accumulated radiation dosage due to continuous image acquisition in CT perfusion (CTP) raised concerns on patient safety and public health. However, low-radiation will lead to increased noise and artifacts which require more sophisticated and time-consuming algorithms for robust estimation. We propose a novel efficient framework using tensor totalvariation (TTV) regularization to achieve both high efficiency and accuracy in deconvolution for low-dose CTP. The method reduces the necessary radiation dose to only 8% of the original level and outperforms the state-of-art algorithms with estimation error reduced by 40%. It also corrects over-estimation of cerebral blood flow (CBF) and under-estimation of mean transit time (MTT), at both normal and reduced sampling rate. An efficient

Tensor total variation (TTV) regularized deconvolution has been proposed for robust low radiation dose CT perfusion. In this paper, we extended TTV algorithm with anisotropic regularization weighting for the temporal and spatial dimension. We evaluated TTV algorithm on synthetic dataset for bolus delay, uniform region variability and contrast preservation, and on clinical dataset for reduced sampling rate with visual and quantitative comparison. The extensive experiments demonstrated promising results of TTV compared to baseline and state-of-art algorithms in low-dose and low sampling rate CTP deconvolution with insensitivity to bolus delay. This work further demonstrates the effectiveness and potential of TTV algorithm's clinical usage for cerebrovascular diseases with significantly reduced radiation exposure and improved patient safety.

Robust deconvolution, the task of estimating hemodynamic parameters from measured spatio-temporal data, is a key problem in computed tomography perfusion. Traditionally, this has been accomplished by solving the inverse problem of the temporal tracer enhancement curves at each voxel inde- pendently. Incorporating spatial contextual information, i.e. information other than the temporal enhancement of the contrast agent, has received significant attention in recent works. Intra-subject contextual information is often exploited to remove the noise and artifacts in the low-dose hemodynamic maps. In this thesis, we take a closer look at the role of inter-subject contextual information in robust deconvolution. Specifically, we explore its importance in three as- pects. First: Informatics acquisition. We show, through synthetic evaluation as well as in-vivo clinical data, that inter-subject similarity provides complimen- tary information to improve the accuracy of cerebral blood flow map estimation and increase the differentiation between normal and deficit tissue. Second: Dis- ease diagnosis. We show that apart from the global learned dictionary for hemo- dynamic maps, the tissue-specific dictionaries can be effectively leveraged for disease diagnosis tasks as well, especially for low-contrast tissue types where the deficits usually occur. Lastly: Treatment plan. We propose a generalized framework with inter-subject context through dictionary learning and sparse representation possible for any hemodynamic parameter estimation, such as blood-brain-barrier permeability. We also extend to include inter-subject context through tensor total variation. The diverse hemodynamic maps provide necessary information for treatment plan decision making. We present results of our approaches on a variety of datasets and clinical tasks, such as uniform regions estimation, contrast preservation, data acquired at low-sampling rate and low radiation dose levels.

Computed tomography perfusion (CTP) is an important functional imaging modality in the evaluation of cerebrovascular diseases, particularly in acute stroke and vasospasm. However, the post-processed parametric maps of blood flow tend to be noisy, especially in low-dose CTP, due to the noisy contrast enhancement profile and the oscillatory nature of the results generated by the current computational methods. In this paper, we propose a robust sparse perfusion deconvolution method (SPD) to estimate cerebral blood flow in CTP performed at low radiation dose. We first build a dictionary from high-dose perfusion maps using online dictionary learning and then perform deconvolution-based hemodynamic parameters estimation on the low-dose CTP data. Our method is validated on clinical data of patients with normal and pathological CBF maps. The results show that we achieve superior performance than existing methods, and potentially improve the differentiation between normal and ischemic tissue in the brain.

Sparse perfusion deconvolution has been recently proposed to effectively improve the image quality and diagnostic accuracy of low-dose perfusion CT by extracting the complementary information from the high-dose perfusion maps to restore the low-dose using a joint spatio-temporal model. However the low-contrast tissue classes where infarct core and ischemic penumbra usually occur in cerebral perfusion CT tend to be over-smoothed, leading to loss of essential biomarkers. In this paper, we extend this line of work by introducing tissuespecific sparse deconvolution to preserve the subtle perfusion information in the low-contrast tissue classes by learning tissue-specific dictionaries for each tissue class, and restore the low-dose perfusion maps by joining the tissue segments reconstructed from the corresponding dictionaries. Extensive validation on clinical datasets of patients with cerebrovascular disease demonstrates the superior performance of our proposed method with the advantage of better differentiation between abnormal and normal tissue in these patients.


Functional imaging serves as an important supplement to anatomical imaging modalities such as MR and CT in modern health care. In perfusion CT (CTP), hemodynamic parameters are derived from the tracking of the first-pass of the contrast bolus entering a tissue region of interest. In practice, however, the post-processed parametric maps tend to be noisy, especially in low-dose CTP, in part due to the noisy contrast enhancement profile and oscillatory nature of results generated by current computational methods. In this paper, we propose a sparsity-based perfusion parameter deconvolution approach that consists of a non-linear processing based on sparsity prior in terms of residue function dictionaries. Our simulated results from numericaldata and experiments in aneurysmal subarachnoid hemorrhage patients with clinical vasospasm show that the algorithm improves the quality and reduces the noise of the perfusion parametric maps in low-dose CTP, compared to state-of-the-art methods

In current computed tomography (CT) examinations, the associated X-ray radiation dose is of significant concern to patients and operators, especially CT perfusion (CTP) imaging that has higher radiation dose due to its cine scanning technique. A simple and cost-effective means to perform the examinations is to lower the milliampere-seconds (mAs) parameter as low as reasonably achievable in data acquisition. However, lowering the mAs parameter will unavoidably increase data noise and degrade CT perfusion maps greatly if no adequate noise control is applied during image reconstruction. To capture the essential dynamics of CT perfusion, a simple spatial-temporal Bayesian method that uses a piecewise parametric model of the residual function is used, and then the model parameters are estimated from a Bayesian formulation of prior smoothness constraints on perfusion parameters. From the fitted residual function, reliable CTP parameter maps are obtained from low dose CT data. The merit of this scheme exists in the combination of analytical piecewise residual function with Bayesian framework using a simpler prior spatial constrain for CT perfusion application. On a dataset of 22 patients, this dynamic spatial-temporal Bayesian model yielded an increase in signal-tonoise-ratio (SNR) of 78% and a decrease in mean-square-error (MSE) of 40% at low dose radiation of 43mA.

A mobile device (160) for medical image analysis is disclosed. The mobile device (160) includes a display (162), a communication module (218), a memory (204) configured to store processor-executable instructions (224) and a processor (202) in communication with the display (162), the communication module (218) and the memory (204). The processor (202) being configured to execute the processor-executable instructions (224) to implement a compression routine to generate a compressed representation of a medical image stored in the memory (204), transmit the compressed representation to a remote device (110) via the communication module (218), receive segmented results from the remote device (110), wherein the segmented results are derived from a reconstruction of the compressed representation generated at the remote device (110), and present, via the display (162), a segmented medical image based on the received segmented results.

We propose a novel approach that applies global optimal tree-metrics graph cuts algorithm on multi-phase contrast enhanced contrast enhanced MRI for liver tumor segmentation. To address the difficulties caused by low contrasted boundaries and high variability in liver tumor segmentation, we first extract a set of features in multi-phase contrast enhanced MRI data and use color-space mapping to reveal spatial-temporal information invisible in MRI intensity images. Then we apply efficient tree-metrics graph cut algorithm on multi-phase contrast enhanced MRI data to obtain global optimal labeling in an unsupervised framework. Finally we use tree-pruning method to reduce the number of available labels for liver tumor segmentation. Experiments on realworld clinical data show encouraging results. This approach can be applied to various medical imaging modalities and organs.

We tackle the challenge of kinship verification using novel feature extraction and selection methods, automatically classifying pairs of face images as “related” or “unrelated” (in terms of kinship). First, we conducted a controlled online search to collect frontal face images of 150 pairs of public figures and celebrities, along with images of their parents or children. Next, we propose and evaluate a set of low-level image features that for use in this classification problem. After selecting the most discriminative inherited facial features, we demonstrate a classification accuracy of 70.67% on a test set of image pairs using K-Nearest-Neighbors. Finally, we present an evaluation of human performance on this problem.

We tackle the problem of brain MRI image segmentation using the tree-metric graph cuts (TM) algorithm, a novel image segmentation algorithm, and introduce a “tree-cutting” method to interpret the labeling returned by the TM algorithm as tissue classification for the input brain MRI image. The approach has three steps: 1) pre-processing, which generates a tree of labels as input to the TM algorithm; 2) a sweep of the TM algorithm, which returns a globally optimal labeling with respect to the tree of labels; 3) post-processing, which involves running the “tree-cutting” method to generate a mapping from labels to tissue classes (GM, WM, CSF), producing a meaningful brain MRI segmentation. The TM algorithm produces a globally optimal labeling on tree metrics in one sweep, unlike conventional methods such as EMS and EM-style geo-cuts, which iterate the expectation maximization algorithm to find hidden patterns and produce only locally optimal labelings. When used with the “tree-cutting” method, the TM algorithm produces brain MRI segmentations that are as good as the Unified Segmentation algorithm used by SPM8, using a much weaker prior. Comparison with the current approaches shows that our method is faster and that our overall segmentation accuracy is better.

Instructor Evaluation: 5.00/5.00, Course Evaluation: 4.75/5.00
Course Objectives: 1. Understand multimodal data mining in the biomedical domain; 2. Understand the concept, approaches, and limitations in analyzing different modalities of biomedical data. 3. Learn to use biomedical data programming libraries and skills to analyze multimodal biomedical data.
The objectives this course are: 1) Develop a proficiency in the use of computer programming (specifically, MATLAB) to analyze biomedical measurements. 2) Develop an understanding of biomedical engineering problems that require quantitative analysis and visualization.
The objectives this course are: 1) Develop a proficiency in the use of computer programming (specifically, MATLAB) to analyze biomedical measurements. 2) Develop an understanding of biomedical engineering problems that require quantitative analysis and visualization.
Instructor Evaluation: 4.63/5.00, Course Evaluation: 4.38/5.00
Course Objectives: 1. Understand multimodal data mining in the biomedical domain; 2. Understand the concept, approaches, and limitations in analyzing different modalities of biomedical data. 3. Learn to use biomedical data programming libraries and skills to analyze multimodal biomedical data.
Instructor Evaluation: 4.92/5.00, Course Evaluation: 4.70/5.00
The objectives this course are: 1) Develop a proficiency in the use of computer programming (specifically, MATLAB) to analyze biomedical measurements. 2) Develop an understanding of biomedical engineering problems that require quantitative analysis and visualization.
Instructor Evaluation: 4.83/5.00, Course Evaluation: 4.53/5.00 (Historical highest for this course)
Course Objectives: 1. Understand multimodal data mining in the biomedical domain; 2. Understand the concept, approaches, and limitations in analyzing different modalities of biomedical data. 3. Learn to use biomedical data programming libraries and skills to analyze multimodal biomedical data.
Instructor Evaluation: 4.87/5.00
The objectives this course are: 1) Develop a proficiency in the use of computer programming (specifically, MATLAB) to analyze biomedical measurements. 2) Develop an understanding of biomedical engineering problems that require quantitative analysis and visualization.
The objectives this course are: 1) Develop a proficiency in the use of computer programming (specifically, MATLAB) to analyze biomedical measurements. 2) Develop an understanding of biomedical engineering problems that require quantitative analysis and visualization.
Data Mining is the nontrivial extraction of implicit, previously unknown, and potentially useful information from data. It has gradually matured as a discipline merging ideas from statistics, machine learning, database and etc. This is an introductory course for junior/senior computer science undergraduate students on the topic of Data Mining. Topics include data mining applications, data preparation, data reduction and various data mining techniques (such as association, clustering, classification, anomaly detection).
Machine learning is concerned with the question of how to make computers learn from experience. The ability to learn is not only central to most aspects of intelligent behavior, but machine learning techniques have become key components of many software systems. For examples, machine learning techniques are used to create spam filters, to analyze customer purchase data, to understand natural language, or to detect fraudulent credit card transactions.
Stolte, Skylar E., Kyle Volle, Aprinda Indahlastari, Alejandro Albizu, Adam J. Woods, Kevin Brink, Matthew Hale, and Ruogu Fang. "DOMINO: Domain-Aware Model Calibration in Medical Image Segmentation." In International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 454-463. Springer, Cham, 2022.
Our SMILE Lab is featured on the Forward & Up video by the College of Engineering!
Dr. Fang is interviewed by ABC Action News in a news segment on the University of Florida's supercomputer, HiPerGator.
UF, NVIDIA partner to speed brain research using AI
University of Florida researchers joined forces with scientists at NVIDIA, UF’s partner in its artificial intelligence initiative, and the OpenACC organization to significantly accelerate brain science as part of the Georgia Tech GPU Hackathon held last month.
Artificial Intelligence Prevents Dementia?
A team at the University of Florida is using targeted transcranial direct current stimulation to save memories. “It’s a weak form of electrical stimulation applied to the scalp. And this weak electric current actually has the ability to alter how the neurons behave,” continued Woods.
UF study shows artificial intelligence’s potential to predict dementia
New research published today shows that a form of artificial intelligence combined with MRI scans of the brain has the potential to predict whether people with a specific type of early memory loss will develop Alzheimer’s disease or other form of dementia.
UF researchers fight dementia using AI technology to develop new treatment
If presented with current models of electrical brain stimulation a decade ago, Dr. Adam J. Woods would have thought of a science-fiction movie plot. Artificial intelligence now pulls the fantasy out of screens and into reality.
UF researchers using artificial intelligence to develop treatment to prevent dementia
UF researchers are developing a method they hope will prevent Alzheimer’s and Dementia. Dr. Ruogu Fang and Dr. Adam Woods are working to personalize brain stimulation treatments to make them as effective as possible.
UF researchers use AI to develop precision dosing for treatment aimed at preventing dementia
UF researchers studying the use of a noninvasive brain stimulation treatment paired with cognitive training have found the therapy holds promise as an effective, drug-free approach for warding off Alzheimer’s disease and other dementias.
Our eyes may provide early warning signs of Alzheimer’s and Parkinson’s
Forget the soul — it turns out the eyes may be the best window to the brain. Changes to the retina may foreshadow Alzheimer’s and Parkinson’s diseases, and researchers say a picture of your eye could assess your future risk of neurodegenerative disease.
Eye Blood Vessels May Diagnose Parkinson's Disease
A simple eye exam combined with powerful artificial intelligence (AI) machine learning technology could provide early detection of Parkinson's disease, according to research being presented at the annual meeting of the Radiological Society of North America.
UF researchers are looking into the eyes of patients to diagnose Parkinson's Disease
With artificial intelligence (AI), researchers have moved toward diagnosing Parkinson’s disease with, essentially, an eye exam. This relatively cheap and non-invasive method could eventually lead to earlier and more accessible diagnoses.
Blood Vessels in the Eye May Diagnose Parkinson's Disease
Using an advanced machine-learning algorithm and fundus eye images, which depict the small blood vessels and more at the back of the eye, investigators are able to classify patients with Parkinson's disease compared against a control group.
Scientists Are Looking Into The Eyes Of Patients To Diagnose Parkinson’s Disease
With artificial intelligence (AI), researchers have moved toward diagnosing Parkinson's disease with, essentially, an eye exam. This relatively cheap and non-invasive method could eventually lead to earlier and more accessible diagnoses.
Eye Exam Could Lead to Early Parkinson’s Disease Diagnosis
A simple eye exam combined with powerful artificial intelligence (AI) machine learning technology could provide early detection of Parkinson’s disease, according to research being presented at the annual meeting of the Radiological Society of North America.
Simple Eye Exam With Powerful Artificial Intelligence Could Lead to Early Parkinson’s Disease Diagnosis
A simple eye exam combined with powerful artificial intelligence (AI) machine learning technology could provide early detection of Parkinson’s disease, according to research being presented at the annual meeting of RSNA.
RSNA 20: AI-Based Eye Exam Could Aid Early Parkinson’s Disease Diagnosis
A simple eye exam combined with powerful artificial intelligence (AI) machine learning technology could provide early detection of Parkinson’s disease, according to research being presented at the annual meeting of the Radiological Society of North America.
Fang selected to ACM's Future of Computer Academy
Dr. Ruogu Fang, incoming assistant professor in the J. Crayton Pruitt Family Department of Biomedical Engineering, has been selected as a member of the Association for Computing Machinery’s (ACM) inaugural Future Computing Academy (FCA).
For Ph.D. applicants interested in working in my lab, you can apply for Ph.D. program in Biomedical Engineering, Electrical and Computer Engineering, Computer Science, or other related programs at UF.
You are welcome to send me an email (ruogu dot fang at bme dot ufl dot edu) with the title "PhD Applicant: YOUR NAME - YOUR UNIVERSITY" and include your CV, transcript, and overview of research experiences before you apply.
If you are already a master student or have received admission to BME/ECE/CISE or related programs at the Universit of Florida, you can contact me via email with the title "MS Applicant: YOUR NAME - YOUR UNIVERSITY" with your CV and transcript with an intention for voluntary research.
We take undergraduate from all years, and prefer students who have a good math and coding background, or at least the strong motivation and interest to build a solid math and coding background.
Please send an email to amylazarte@ufl.edu and me with the title "Undergraduate Applicant: YOUR NAME - YOUR YEAR IN THE PROGRAM (e.g., sophomore, junior)" including your CV, transcript, and a brief research statement of why you want to join SMILE lab as an undergraduate researcher, what related research projects you have been involved, and why you think you will be a good undergraduate researcher in our lab.Prerequisite: Complete Coursera Machine Learning course by Andrew Ng before you can officially join the lab. (Free)
Co-requisite: Students who have joined our lab (including PhD, MS, and UG) are strongly encouraged to complete the following courses online during their time in the lab:
All students that meet these requirements are welcome to apply! We are particularly interested in broadening participation of underrepresented groups in STEM, and women and minorities are especially encouraged to apply. We also aim to provide research experiences for students who have limited exposure and opportunities to participate in research.
I would be happy to talk to you if you need my assistance in your research, have intention for collaboration, or need technology translation for your company. Also, welcome to join my SMILE research group. Email is the best way to get in touch with me. Please feel free to contact me!
J287 Biomedical Science Building
University of Florida
1275 Center Dr.
Gainesville, FL, 32611.